BtaEX0017283 @ bosTau6
Exon Skipping
Gene
ENSBTAG00000005726 | HNRNPA2B1
Description
Bos taurus heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1), mRNA. [Source:RefSeq mRNA;Acc:NM_001045975]
Coordinates
chr4:70198053-70200712:+
Coord C1 exon
chr4:70198053-70198191
Coord A exon
chr4:70200469-70200504
Coord C2 exon
chr4:70200602-70200712
Length
36 bp
Sequences
Splice sites
3' ss Seq
AGTTTTTTTTTTCCCCCCAGAAA
3' ss Score
10.14
5' ss Seq
AAGGTACTC
5' ss Score
7.46
Exon sequences
Seq C1 exon
CCAGCGGTGTTGTTTCTCGAGCAGCGGCAGTTCTCACTACAGCGCCAGGACGAGTCCGGTTCGTGTTCGTCCGCGGAGATCTCTCTCATCTCGCTCGGCTGCGGGAAATCGGGCTGAAGCGACTGAGTCCGCGATGGAG
Seq A exon
AAAACTTTAGAAACTGTTCCTTTGGAGAGGAAAAAG
Seq C2 exon
AGAGAAAAGGAACAATTCCGTAAACTCTTTATTGGTGGCTTGAGCTTTGAAACTACAGAAGAAAGTTTGAGGAACTACTACGAGCAATGGGGAAAACTTACAGATTGTGTG
VastDB Features
Vast-tools module Information
Secondary ID
ENSBTAG00000005726_CASSETTE1
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.167 A=0.833 C2=0.000
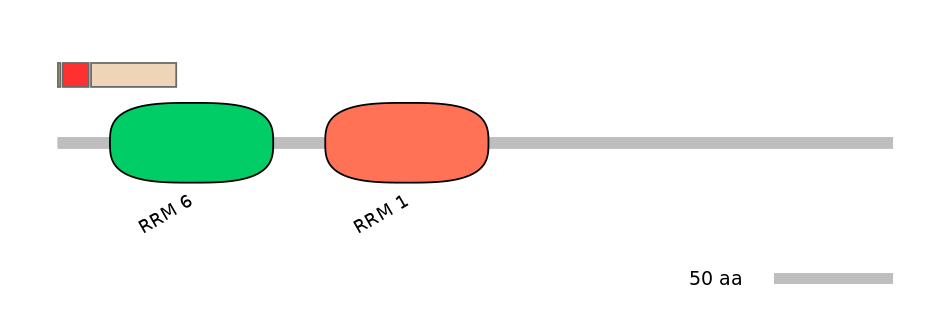
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
PF142591=RRM_6=PU(40.0=75.7)
Main Inclusion Isoform:
ENSBTAT00000007527fB3267
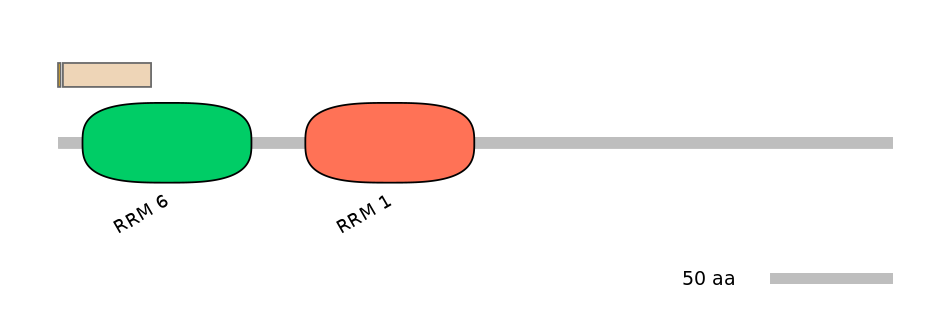
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ATCTCTCTCATCTCGCTCGGC
R:
AGTTTCAAAGCTCAAGCCACCA
Band lengths:
115-151
Functional annotations
There are 1 annotated functions for this event
PMID: 20406423
The inclusion of exon 2 (HsaEX0030279) and exon 9 (HsaEX0030282) generates a predominantly nuclear form, skipping of the exons generates cytosolic forms. The study shows, using isoform-specific antibodies and isoform-specific green fluorescent protein (GFP)-fusion expression constructs, that A2b is the predominant cytoplasmic isoform in neural cells, suggesting that it may play a key role in mRNA trafficking. The differential subcellular distribution patterns of the individual isoforms are determined by the presence or absence of alternative exons that also affect their dynamic behavior in different cellular compartments, as measured by fluorescence correlation spectroscopy. Expression of A2b is also differentially regulated with age, species and cellular development. Furthermore, coinjection of isoform-specific antibodies and labeled RNA into live oligodendrocytes shows that the assembly of RNA granules is impaired by blockade of A2b function. These findings suggest that neural cells modulate mRNA trafficking by regulating alternative splicing of hnRNP A2/B1 and controlling expression levels of A2b, which may be the predominant mediator of cytoplasmic-trafficking functions. B1: inclusion of both. A2: skipping of ex 2. B1b: skipping of ex 9. A2b: skipping of both. Not well in VTS in hg38 nor mm10.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development