BtaEX0019886 @ bosTau6
Exon Skipping
Gene
ENSBTAG00000008817 | LAMA4
Description
Bos taurus laminin, alpha 4 (LAMA4), mRNA. [Source:RefSeq mRNA;Acc:NM_001205965]
Coordinates
chr9:38790867-38794022:+
Coord C1 exon
chr9:38790867-38791031
Coord A exon
chr9:38793245-38793256
Coord C2 exon
chr9:38793869-38794022
Length
12 bp
Sequences
Splice sites
3' ss Seq
TCTTGTCTTTTTTATGATAGGCA
3' ss Score
7.62
5' ss Seq
CAGGTGAGT
5' ss Score
10.67
Exon sequences
Seq C1 exon
GTATGAATTGATAGTAGATAAAAGCAGACTTGGGAGTAAGAACCCTACCAAAGGGAAAGTGGAGCAGACCCAAGCAGATGAAAAGAAGTTCTACTTCGGCGGCTCACCCATCACTCCCCAGTATGCTAATTTCACCGGATGTATAAGTAATGCCTACTTTACCAG
Seq A exon
GCATCATGACAG
Seq C2 exon
GTTGGATAGAGATGTAGAGGTCGAAGATTTCCAGCGGTATTCTGAAAAGGTCCACACTTCTCTTTATGAGTGTCCCATTGAGTCTTCACCATTGTTTCTCCTTCACAAAAAAGGAAGAAATTCCTCAAAGCCTAAAACAAATCAGAATAAAAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSBTAG00000008817-MICROEX1
Average complexity
MIC_S
Mappability confidence:
NA
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.143 A=0.000 C2=0.317
Domain overlap (PFAM):
C1:
PF0221019=Laminin_G_2=FE(47.4=100)
A:
PF0221019=Laminin_G_2=PD(0.9=20.0)
C2:
PF0221019=Laminin_G_2=PD(0.1=0.0)
Main Inclusion Isoform:
ENSBTAT00000011612fB3768
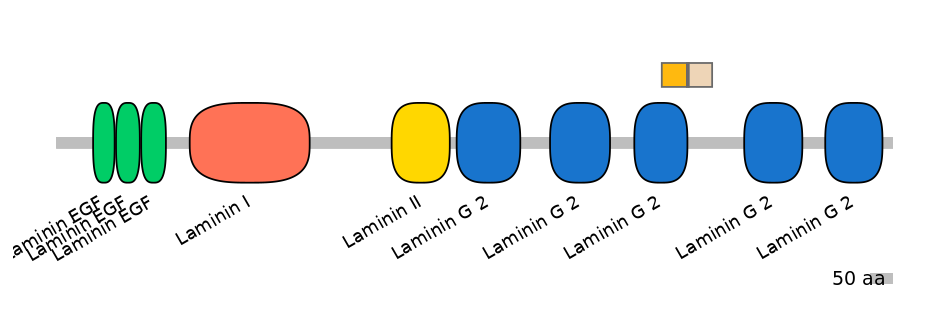
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CCCATCACTCCCCAGTATGCT
R:
CCGCTGGAAATCTTCGACCTC
Band lengths:
96-108
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development