DmeEX0000205 @ dm6
Exon Skipping
Gene
FBgn0261788 | Ank2
Description
The gene Ankyrin 2 is referred to in FlyBase by the symbol DmelAnk2 (CG42734, FBgn0261788). It is a protein_coding_gene from Dmel. It has 21 annotated transcripts and 21 polypeptides (20 unique). Gene sequence location is 3L:7655389..7718395. Its molecular function is unknown. It is involved in the biological process described with 9 unique terms, many of which group under: nervous system process; system process; positive regulation of multicellular organismal process; regulation of trans-synaptic signaling; organelle organization. 41 alleles are reported. The phenotypes of these alleles manifest in: cellular anatomical entity; cell junction; synapse; cell component; dendritic tree. The phenotypic classes of alleles include: stress response defective; increased mortality; phenotype; neuroanatomy defective.
Coordinates
chr3L:7656375-7663324:-
Coord C1 exon
chr3L:7663259-7663324
Coord A exon
chr3L:7662471-7662890
Coord C2 exon
chr3L:7656375-7661506
Length
420 bp
Sequences
Splice sites
3' ss Seq
ACTGTGCAATTGTCTTCCAGGTG
3' ss Score
11.12
5' ss Seq
AAGGTAACA
5' ss Score
8.92
Exon sequences
Seq C1 exon
AAACCCTTCACGAAAGAAAAGTAGAACTATCCAATCTGGATACCGAAAAGATCTCACATGAAATTG
Seq A exon
GTGATGTTAAAGTTGGTGGTGGTGATGATGATAGTGATGGCGATAGTGGAGATTTTGTGGCACGCATTCGCGAGGAAGCCAATATTCTGGTGGATCGCGTACTTGAAGAAAGTGTCGAGATCATTGAGAGTCAAGGCCATCAGGCCAATGGCCACGAGATCGCTAAACTGGACTACAACTCAGAAAGTGAGAATTATGCCATCACATGCGATGATATCGGCGGCGTCGATGTTATCAAATCGCCCACCATTGAATCCATGTCGGGCAAGTCGTTTGACGACAACATGAGCTTCACCGACGAGCATATACATTTACCCTTGCATGCCCAGAACACGAGCACCACCATGACAACGACAATTACCACTCAGTTCCAACAGCAGCAGTCCCAGTCTGCTGTCTCAAGTACATCGGCGGCAAAAG
Seq C2 exon
AAACAGGCGGCGATTTACCAAACTCAAGTGAAACCGATACCACTCCCGTACCACCGTCGAAAGAATCGGAAAGCAAAACCGAAACTGAGCCGACGGTAGTAACCAATGCATTCACTACCACCAGCCACGATACACCCACACCAAAACCAAGAGTACTCAAAGCAAGCTCCCCCGTCTCCGAAGGCACTGATAGCACTACCATTCCTGGCCCTGTGCCCAAGCCCGAAATACCGATTGATATTAGTTTCGATGCCAACGCACCGGTACCAAAGCCGCGTGCCCCTATTAAGCACACGGAACCTTTCGAAAACCTGGCTGCCCTAGCTGAACAGTCCGATAGCATTAGTCTGAGGAGTTTTGAGCTCACAGAGGACATTTCCAAAACGGACGAGCCCTCCACAGAGCTCTCCATTCGATCCCAGCTTCGGGACATAGTCACCACCACCACTGTTTCGGAGGATCACACCGAAAGCTTTGAGCCAACGAGTGAGATTTCTGTC
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0261788_MULTIEX1-1/3=C1-3
Average complexity
C2
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.524 A=0.713 C2=0.821
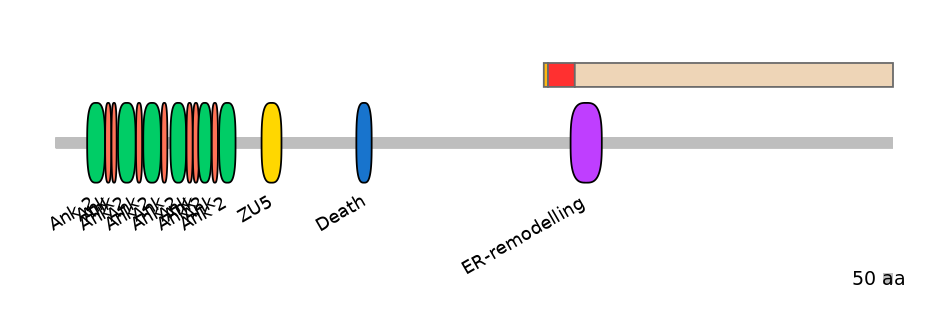
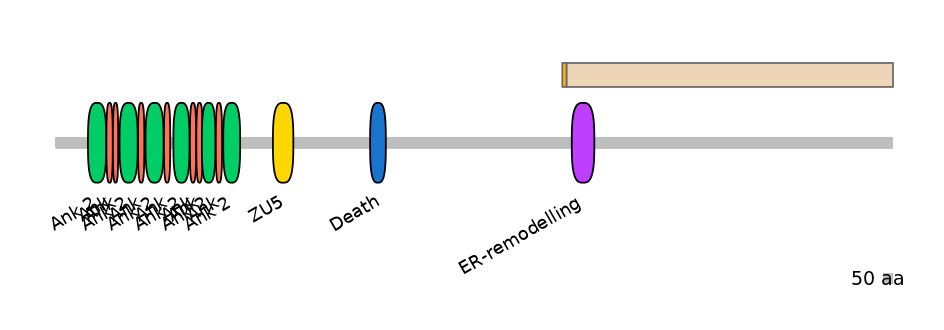
Domain overlap (PFAM):
C1:
NO
A:
PF147551=ER-remodelling=PU(13.4=15.6)
C2:
PF147551=ER-remodelling=PD(86.0=8.5)
Main Inclusion Isoform:
FBpp0306185
Main Skipping Isoform:
FBpp0292243
Other Inclusion Isoforms:
FBpp0292230, FBpp0292231, FBpp0292232, FBpp0292233, FBpp0292234, FBpp0292244, FBpp0310628
Other Skipping Isoforms:
FBpp0292236, FBpp0292240, FBpp0292242, FBpp0310629
Associated events
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TCTGGATACCGAAAAGATCTCACA
R:
GCAGCCAGGTTTTCGAAAGGT
Band lengths:
351-771
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)