DmeEX0002724 @ dm6
Exon Skipping
Gene
FBgn0267431 | CG45784
Description
The gene Myosin 81F is referred to in FlyBase by the symbol DmelMyo81F (CG45784, FBgn0267431). It is a protein_coding_gene from Dmel. It has 2 annotated transcripts and 2 polypeptides (1 unique). Gene sequence location is 3R:567076..2532932. Its molecular function is described by: actin-dependent ATPase activity; ATP binding; microfilament motor activity; actin filament binding. It is involved in the biological process described with: actin filament organization; signal transduction; sensory organ development; vesicle transport along actin filament; sensory perception of sound. 6 alleles are reported. No phenotypic data is available. The phenotypic class of alleles includes: viable.
Coordinates
chr3R:1191610-1342317:+
Coord C1 exon
chr3R:1191610-1191975
Coord A exon
chr3R:1297134-1297392
Coord C2 exon
chr3R:1342196-1342317
Length
259 bp
Sequences
Splice sites
3' ss Seq
TGCACTTTTATTTGGAATAGGTA
3' ss Score
6.84
5' ss Seq
CCAGTGAGT
5' ss Score
8.28
Exon sequences
Seq C1 exon
TATGAACATGAAGGAATATACTTTAATCATGTGGAGTTTACAGATAATACTCCTTGCTTGGAAATATTAGAAAAACCTCCGCGATGTGTGTTCAAGTTACTGACAGAACAGTGTCATATGCCACAAGGCTCTGATTTGGCTTTTTTAAGTAATATGCACACTGAGTTCGAGTCACACCCAAATTACATTAAAGGATCTGATCGTCGTCATTGGGAAACCGAGTTTGGAATAAAACATTATGCAGGTCTTGTAATCTACGCTGTGGGCGGATTTGTGGAAAAAAATCGAGATGTACAACAAGATGTAATGTTTGAATACATGAGCTACAGCAAAGACGTGTTTATAAAAGATTTATCAACATACCAG
Seq A exon
GTACTGTTACCAAATATTTTGTTAAATCGCCCAGTGGCGGAGATTTTGGAGTCGTCGAAGGTTTCCCAATGGAGATGGGTGCCTACAGCTGGCAATGCGGCTGATGATGCGACGCGGTCGCAGAAAGGAGTCGACCTTAGCCAGGAATCAAGGTGGCTAAGAGGACCTGCATTTTTGAGGCAGCCAGCAGCCAGCTGGCCGGGGCCAGAGGAAGGAACTGAGCGTGTTCCAGATGCCCCTGATGAAGAAGAGATGCCCA
Seq C2 exon
GATATCCAATTCATCAATTCAAGTTATCCACGAGGATCATCGAAATCAAAATGTACTGTAAGCGACAATTTTCGTAATCAGTTACAGTCCCTCATAGATGTTCTCCAAAATACAAAGCCTTG
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0267431-'10-11,'10-10,11-11
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
In the CDS, with uncertain impact
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.333 C2=0.030
Domain overlap (PFAM):
C1:
PF0006316=Myosin_head=FE(18.2=100)
A:
PF0006316=Myosin_head=PD(7.7=50.6)
C2:
PF0006316=Myosin_head=FE(6.0=100)
Main Inclusion Isoform:
FBtr0346770fB332
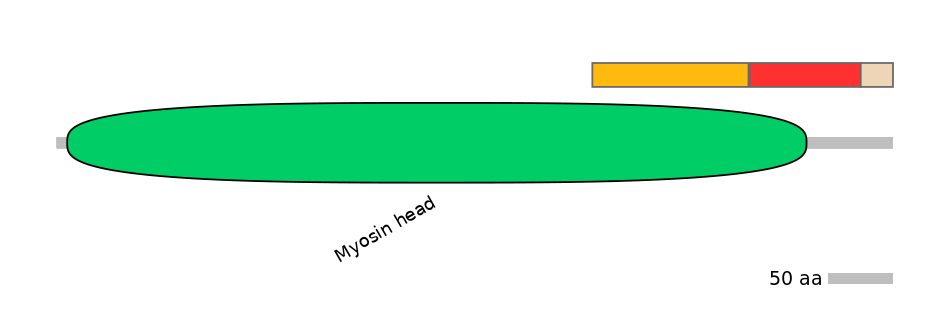
Main Skipping Isoform:
FBpp0312365
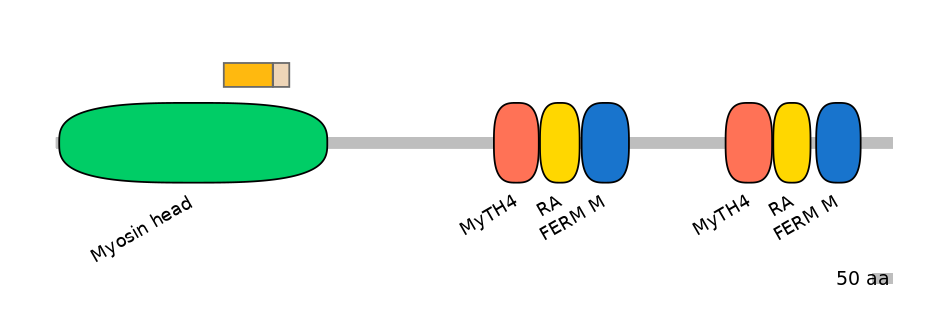
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CCACAAGGCTCTGATTTGGCT
R:
TGATTTCGATGATCCTCGTGG
Band lengths:
294-553
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)