DmeEX0004375 @ dm6
Exon Skipping
Gene
FBgn0266084 | Fhos
Description
The gene Formin homology 2 domain containing is referred to in FlyBase by the symbol DmelFhos (CG42610, FBgn0266084). It is a protein_coding_gene from Dmel. It has 10 annotated transcripts and 10 polypeptides (8 unique). Gene sequence location is 3L:8753619..8798719. Its molecular function is described by: actin filament binding. It is involved in the biological process described with 12 unique terms, many of which group under: anatomical structure morphogenesis; epithelium development; multicellular organismal process; positive regulation of cellular component movement; programmed cell death involved in cell development. 43 alleles are reported. The phenotypes of these alleles manifest in: intracellular non-membrane-bounded organelle; multicellular structure; intracellular organelle; cytoskeleton; intracellular. The phenotypic classes of alleles include: increased mortality during development; stress response defective; phenotype; fertile.
Coordinates
chr3L:8766442-8782657:+
Coord C1 exon
chr3L:8766442-8766629
Coord A exon
chr3L:8776997-8777317
Coord C2 exon
chr3L:8782548-8782657
Length
321 bp
Sequences
Splice sites
3' ss Seq
ATGGCAAAAATCCATTTCAGTGT
3' ss Score
3.03
5' ss Seq
AGCGTAAGA
5' ss Score
6.21
Exon sequences
Seq C1 exon
TCAGTTGCCTTGCGAACGCGCTCAAGTTCAGTCGCTTCGATTTTGTCGCGGGTGAAACAACCACCGTGGAATCAAAAACCAAACTTCGCTCAGCTGCGCGCGTCGCGTTGCCCTGTGTGTGTGCTTTTGTTGTTGTTGAGGGCAACGTTTAAAATATATTTAAACTGCCTTAGTTGCTACAACAATTG
Seq A exon
TGTTCGTTCAAGTGTTCGAGTGAAAATGTAAATAAATCATAGAGCGAGTGAGAGAGAGAAAATCGCGAAAAGTAAATAGACCTCGAATTAAGTAAGACAAATGTGCAAAAGTGATTGAAAGCCGCGTCGAAAATGATTGTGAAAATGGAGCCGGACGAACTGATCACATTCCGTGTCCAGTATCTGGTGGACAGCGATCCGCTCTCGGCGTGCACCGCCTTCCCGATCCCGGTGCGGGCGCCCACCTACGCCTTCGCCACCACCATGCCGCTGGCCAATCAGCTGGGCACAATCCTGCGGCTGCTGAATGCACCACAAAGC
Seq C2 exon
ATCGACGATGCAGCCATTCAGGTGTACAAGGATGGCGACTACGGAGCCTACTTGGATCTGGAGTCATCGCTGGCCGAACAGGCCGAGGAAATCGAGGGAGTCAACGTCAG
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0266084-'6-6,'6-5,12-6
Average complexity
C3
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref, Alt. ATG (>10 exons))
No structure available
Features
Disorder rate (Iupred):
C1=NA A=0.000 C2=0.054
Domain overlap (PFAM):
C1:
NA
A:
NO
C2:
NO
Main Inclusion Isoform:
FBpp0291739
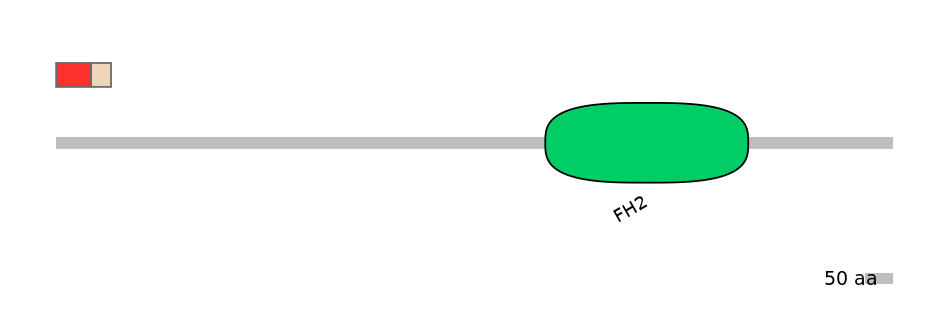
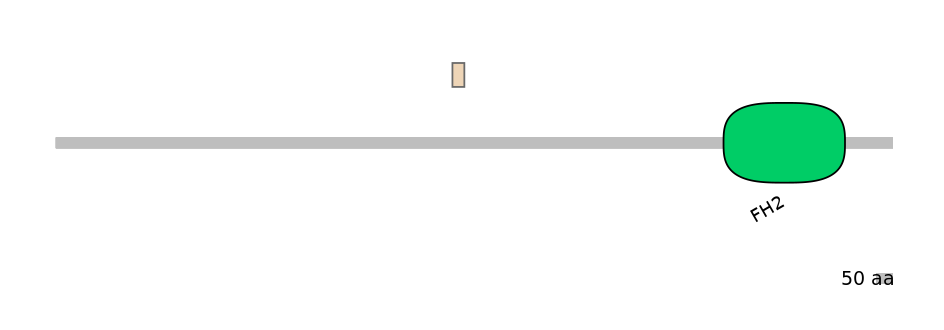
Main Skipping Isoform:
FBtr0333888fB502
Other Inclusion Isoforms:
FBpp0291295, FBpp0291296, FBpp0291740, FBpp0297586
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg19)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Chicken
(galGal3)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TCAGTTGCCTTGCGAACGC
R:
ACGTTGACTCCCTCGATTTCC
Band lengths:
295-616
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)