DmeEX6004937 @ dm6
Exon Skipping
Gene
FBgn0016920 | nompC
Description
The gene no mechanoreceptor potential C is referred to in FlyBase by the symbol Dmel
ompC (CG11020, FBgn0016920). It is a protein_coding_gene from Dmel. It has 6 annotated transcripts and 6 polypeptides (5 unique). Gene sequence location is 2L:5346200..5365760. Its molecular function is described by: cation channel activity; mechanosensitive ion channel activity; calcium channel activity; ion channel activity; ankyrin binding. It is involved in the biological process described with 13 unique terms, many of which group under: response to abiotic stimulus; multicellular organismal process; nervous system process; system process; response to external stimulus. 31 alleles are reported. The phenotypes of these alleles manifest in: adult prothoracic segment; mechanosensory chaeta; cell projection; larval dorsal multidendritic neuron ddaA; adult neuron. The phenotypic classes of alleles include: sensory perception defective; behavior defective; some die during larval stage; phenotype. Summary of modENCODE Temporal Expression Profile: Temporal profile ranges from a peak of moderate expression to a trough of extremely low expression. Peak expression observed within 18-24 hour embryonic stages.
Coordinates
chr2L:5363656-5365382:+
Coord C1 exon
chr2L:5363656-5363784
Coord A exon
chr2L:5364309-5364417
Coord C2 exon
chr2L:5364774-5365382
Length
109 bp
Sequences
Splice sites
3' ss Seq
TCAAAAAACACGCGCCTCAGGCA
3' ss Score
-0.15
5' ss Seq
AAGGTGAGA
5' ss Score
8.68
Exon sequences
Seq C1 exon
GCCCAATCCGACATCGAGTGGAAATTTGGCTTGTCCAAGCTTATACGCAATATGCATCGCACCACAACAGCGCCATCGCCGCTTAATTTAGTTACCACCTGGTTTATGTGGATCGTCGAGAAGGTCAAG
Seq A exon
GCACGCATGAAGAAAAAGAAGCGTCCAAGTCTGGTTCAGATGATGGGAATACGTCAGGCCAGTCCGCGTACCAAAGCCGGCGCCAAGTGGCTGTCGAAGATCAAGAAAG
Seq C2 exon
ACTCAGTGGCCCTGTCGCAGGTCCATCTATCGCCTCTGGGATCACAGGCGAGCTTCTCGCAGGCCAATCAGAATCGCATCGAGAACGTGGCCGACTGGGAGGCGATTGCCAAAAAGTACCGGGCACTGGTTGGCGACGAGGAGGGTGGATCGCTCAAGGACTCGGATGCGGAGAGTGGATCGCAGGAGGGTAGCGGAGGACAACAGCCACCGGCACAGGTGGGCAGACGAGCCATCAAGGCCACCCTGGCAGACACTACAAAATAGACACACAGAAATGACACAGAAAAAACAGAAAAACAGCTTCGGATGCTTAATTAACTACGTTTTGATTGCAGGTCTAAGCTTCATCTATCTCTTCAAACTATCCTTCCTGACTATCTCTATCTCTTCTCGACTATCCAAGCGTCTGTCCTTCTGTAATTCTAAGATCTAACTCTAAGAAACTCTATCCGTAAGCTGCACCTTGGGTATGGTTTTCTCAGACTCTGGAACCCACTT
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0016920-'17-16,'17-15,18-16=AN
Average complexity
A_S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref, Alt. Stop)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.053 C2=0.518
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
NO
Main Inclusion Isoform:
FBpp0289289
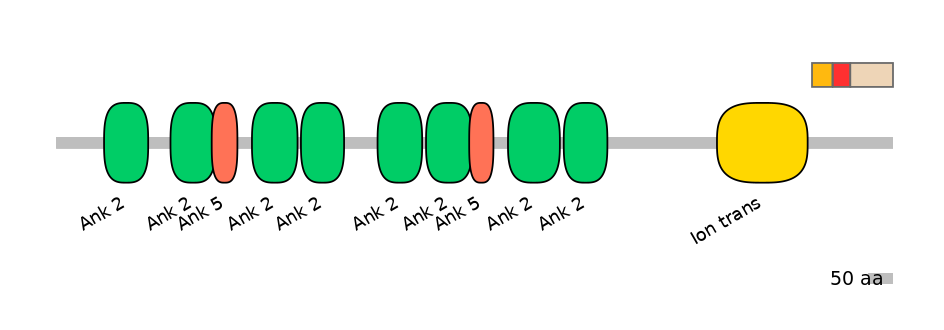
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
FBpp0293383, FBpp0309913
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TACGCAATATGCATCGCACCA
R:
CATCCGAGTCCTTGAGCGATC
Band lengths:
254-363
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)