DmeINT0015486 @ dm6
Intron Retention
Gene
FBgn0037717 | CG8301
Description
This gene is referred to in FlyBase by the symbol DmelCG8301 (FBgn0037717). It is a protein_coding_gene from Dmel. It has 2 annotated transcripts and 2 polypeptides (1 unique). Gene sequence location is 3R:9581306..9586648. Its molecular function is described by: nucleic acid binding; zinc ion binding. It is involved in the biological process described with: . 12 alleles are reported. No phenotypic data is available. The phenotypic classes of alleles include: viable; fertile. Summary of modENCODE Temporal Expression Profile: Temporal profile ranges from a peak of moderately high expression to a trough of very low expression. Peak expression observed within 00-06 hour embryonic stages, during late pupal stages.
Coordinates
chr3R:9585749-9586648:+
Coord C1 exon
chr3R:9585749-9585898
Coord A exon
chr3R:9585899-9585955
Coord C2 exon
chr3R:9585956-9586648
Length
57 bp
Sequences
Splice sites
5' ss Seq
AGAGTATGT
5' ss Score
5.66
3' ss Seq
TAGATGAACATCACTTCCAGCTC
3' ss Score
2.28
Exon sequences
Seq C1 exon
TCTTGGAGAAGAACTTCAAGTGCAACGTTTGCGGTCGAGCCTATCTGTTTGAGAAGTCATTGCGGCTGCATCAGCGAACTCACACGGGCAAAACCTACTACAAATGCGATCTCTGTCAGGAAAGATTTGTCACGCACATTAAGCTCAAGA
Seq A exon
GTATGTATGCGAAATCACTCAATGATGCCCTTCCATATAGATGAACATCACTTCCAG
Seq C2 exon
CTCACATGCAAAAGGCACACGCCGCCTCTCAGCCGCACTCGGCGGACCAGCCTCTGGACGATCTGATCAACATTGTGATTTCCTAACCAAGTGGAACAGCAAACAGCATAGATTTCTGTTGTAGCCAAGCACAGTGCAGAAACTGCAATGCATTGGAGCGTAGCTCCCGAAACTCAAGCTTACATAGACCATATACATACATGTACCCTCCATCCAATTACACTTACTAGACCATATGCACACACGCAGCGATCTCTGGAACGTGAAAGTGCTTAGAGCTTTATTTAGTGGATATTCCGTAGGGCATAAACCATATAGCTATATCTCTGTCCATTTCTTCGATCGGTCCCTAAGGCACTCCAGCCGCATATCTCTTTCTAACTCTGAGCCATATTAATCTCACAGAACGCCTTTAAACACTACCAGCACATACATACATATATCCATTTGACCACTTAGCGATAAGAAATAAGCAATGAGAACATACAAGATGTTATACGAGCAATCTAAGCTTCGCGGCGAACTTTTAATGCTCAAGAAATGCATTTATTTCCAGATGAAACCGATAAGTATTTACAATAATTACCGAAAATGTATAAGAATATTTAAAATATCAATATACTATGGATTTTATTATGTGTAGTTGCAGATATATTTACCAAATTCATGTTCCGAATAAACATTCATGTGACC
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0037717:FBtr0082073:5
Average complexity
IR
Mappability confidence:
NA
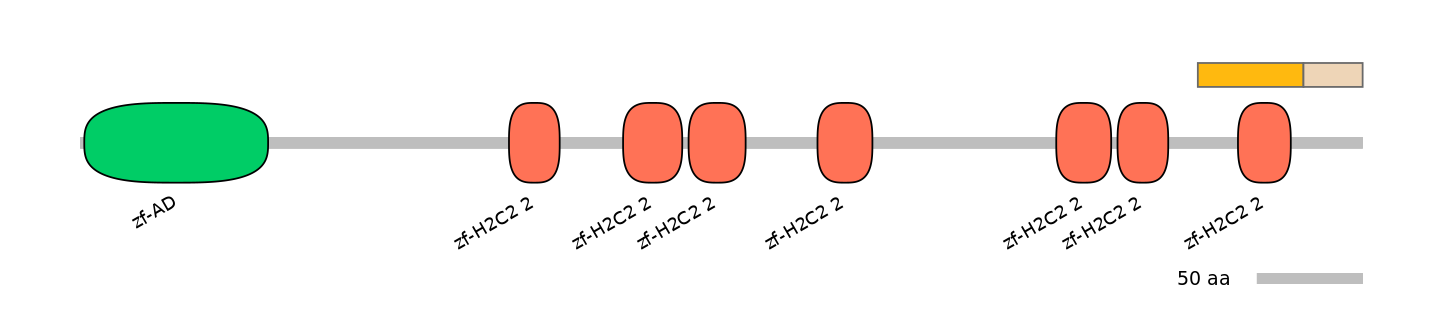
Protein Impact
Alternative protein isoforms
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=NA C2=0.357
Domain overlap (PFAM):
C1:
PF051298=Elf1=FE(73.5=100),PF134651=zf-H2C2_2=WD(100=51.0)
A:
NA
C2:
PF051298=Elf1=PD(7.4=17.2)
Main Inclusion Isoform:
NA
Main Skipping Isoform:
FBpp0081551
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ACTCACACGGGCAAAACCTAC
R:
TGATCAGATCGTCCAGAGGCT
Band lengths:
142-199
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)