DmeINT0030056 @ dm6
Intron Retention
Gene
FBgn0003887 | betaTub56D
Description
NA
Coordinates
chr2R:19447319-19449599:-
Coord C1 exon
chr2R:19449327-19449599
Coord A exon
chr2R:19448747-19449326
Coord C2 exon
chr2R:19447319-19448746
Length
580 bp
Sequences
Splice sites
5' ss Seq
TGTGTATGT
5' ss Score
3.3
3' ss Seq
CTTCCATTTCTCATTTGCAGTTC
3' ss Score
10.77
Exon sequences
Seq C1 exon
ATAGTAGTTATACAGCTCAGTTCCAGCCCACGCCCGAAGATTCGAGTAAGATCGGTCGGCGGAACGCAATGACAGCTGCGCCTCGAGATCCGAAACGAAAACCCACGCCAGATGACGTTATCGTTGGTGTGGCGATGGTGCTGCTGGTCGTGGTGGTGGAAACGCACCAAAAATGGCAGCGAATAAGTTATGTTAATCGGAGCTCGGCCCATACACAGAGTTTATGATGGCGTACATGGGTGGCCTAAACCACGTAGCCGAGCCCAGACATGT
Seq A exon
GTATGTACATCGGCTGCTTCTTGTGCCCATTTATTCCAATCAATAAAGAGTTATATATAGACTGACTTTCCATAACCCATCACACATTTTGTTCGAGAAGGTTTCTGTATCGAAGTTATCTGTATAAGTTAAGAGAAAAACAAGAAAATAAGATTGCCTTTCACATGCATCAACTTTCACTGGGTATGGAAGAAAAATTTTAAGCAAATTACTTGCAGAATTGGAAATGAGTTTCAGTGATTAGTAAAAGTTTTCTTCAACTGCAGTACTCTTGTGCAAATAAAAGTTACCGTTGCTTAAGCATAAACTTCTTTTGTGATTTATGTGAAGCAGTCAAATAAATACTAGCTGTATATATGTATGCAGCAGATGGTCTAGGCTTTGAGTAATTTTACTACCATTGATTTTCTGTGATGACTCATTGGCCAAGTCATTCATACATTTTCTTAAGACTGAGTTAGAAAACACCCCCTAGTAATCGATTTTCCTGCTGACTCATGTTCACAAGAAATGCTGTACAGCACCTGCAAGTCCTAATCGCACTAATCCCAAATTCTTTACTTCCATTTCTCATTTGCAG
Seq C2 exon
TTCTGGGAGATCATCTCCGATGAGCATGGCATCGATGCCACCGGCGCCTACCACGGTGACAGCGACCTGCAGCTGGAGCGCATCAATGTGTACTACAATGAGGCGTCCGGTGGCAAGTACGTGCCCCGCGCTGTCCTTGTCGATCTGGAGCCCGGCACCATGGACTCTGTGCGATCGGGACCTTTCGGCCAGATCTTCAGGCCCGACAACTTTGTGTTCGGCCAGTCGGGTGCCGGCAACAACTGGGCCAAGGGTCATTACACAGAGGGTGCTGAGCTGGTGGACTCAGTGCTCGATGTTGTCCGCAAGGAGGCCGAATCCTGCGACTGCCTGCAAGGCTTCCAACTCACACACTCCCTTGGCGGCGGCACTGGCTCCGGTATGGGAACCCTGCTGATTTCCAAGATCCGCGAGGAGTACCCCGACAGGATCATGAACACATACTCGGTTGTGCCGTCGCCCAAGGTGTCGGACACCGTTGTGGAGCCCTACAACGCCACCCTGTCGGTGCACCAGCTGGTCGAGAACACGGACGAGACCTACTGCATCGACAACGAGGCTCTCTACGATATCTGCTTCCGCACCCTCAAGCTGACGACACCCACATACGGTGACCTGAACCATCTTGTCTCCCTGACCATGTCCGGCGTAACCACCTGCCTCCGATTCCCCGGCCAACTGAACGCTGATCTCCGCAAGCTGGCCGTCAACATGGTGCCCTTCCCACGTCTTCACTTCTTCATGCCCGGCTTCGCTCCCCTGACCTCCCGAGGATCCCAGCAGTACCGCGCCCTCACCGTGCCCGAGCTGACCCAGCAGATGTTCGATGCCAAGAACATGATGGCCGCCTGCGACCCACGCCACGGACGCTACCTTACCGTCGCCGCCATCTTCCGTGGACGCATGTCCATGAAGGAGGTCGACGAGCAGATGCTGAACATCCAGAACAAGAACAGCTCCTACTTCGTCGAATGGATCCCCAACAACGTGAAGACCGCCGTGTGCGACATCCCGCCCCGTGGTCTGAAGATGTCCGCCACCTTCATCGGCAACTCCACTGCCATCCAGGAGCTGTTCAAGCGCATCTCCGAGCAGTTCACCGCTATGTTCAGGCGCAAGGCTTTCTTGCATTGGTACACCGGCGAGGGCATGGACGAGATGGAGTTCACCGAGGCCGAGAGCAACATGAACGATCTGGTGTCCGAGTACCAGCAGTACCAGGAGGCCACCGCCGACGAGGACGCCGAGTTCGAGGAGGAGCAGGAGGCTGAGGTCGACGAGAACTAAATTCGAATCGGAAATCAATCGAATTCTCGATCAATGTTTTGCGAAAAAAAATGAAACGAGAACAAATGAATTCAGTTGTAAAAGATTTGTTCTCTTAACAACGTAATCAATAAAGTCGGATCCAATTTTGTTCACAAGTCT
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0003887:FBtr0086537:1
Average complexity
IR
Mappability confidence:
NA
Protein Impact
Alternative protein isoforms
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=NA C2=0.086
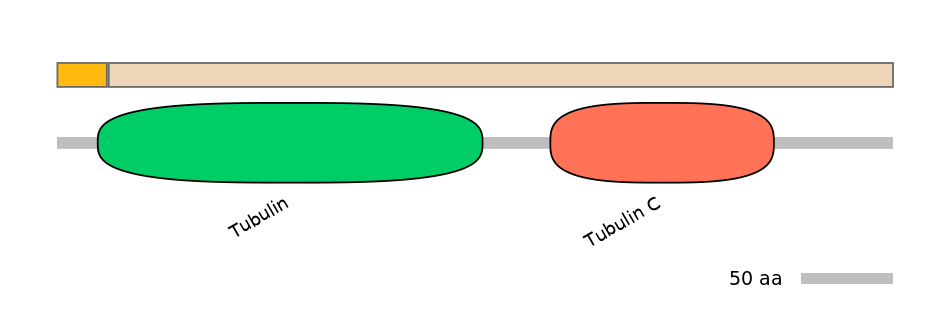
Domain overlap (PFAM):
C1:
PF0009120=Tubulin=PU(2.4=17.9)
A:
NA
C2:
PF0009120=Tubulin=PD(91.9=47.6),PF0395312=Tubulin_C=WD(100=28.7)
Main Inclusion Isoform:
NA
Main Skipping Isoform:
FBpp0085721
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TTAATCGGAGCTCGGCCCATA
R:
GTGTAATGACCCTTGGCCCAG
Band lengths:
344-924
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)