DmeINT0034634 @ dm6
Intron Retention
Gene
FBgn0002939 | ninaD
Description
The gene neither inactivation nor afterpotential D is referred to in FlyBase by the symbol Dmel
inaD (CG31783, FBgn0002939). It is a protein_coding_gene from Dmel. It has 3 annotated transcripts and 3 polypeptides (2 unique). Gene sequence location is 2L:18081628..18083608. Its molecular function is described by: scavenger receptor activity. It is involved in the biological process described with: phototransduction, visible light; carotenoid transport; rhodopsin biosynthetic process; phototransduction, UV; phototransduction. 12 alleles are reported. The phenotype of these alleles manifest in: photoreceptor cell. The phenotypic classes of alleles include: viable; neurophysiology defective; lethal; lethal - all die during embryonic stage. Summary of modENCODE Temporal Expression Profile: Temporal profile ranges from a peak of moderately high expression to a trough of extremely low expression. Peak expression observed during late larval stages, in adult male stages.
Coordinates
chr2L:18082224-18083189:+
Coord C1 exon
chr2L:18082224-18083035
Coord A exon
chr2L:18083036-18083098
Coord C2 exon
chr2L:18083099-18083189
Length
63 bp
Sequences
Splice sites
5' ss Seq
AGAGTAATT
5' ss Score
1.61
3' ss Seq
ATAAACACATTTTTTCCTAGCAT
3' ss Score
4.08
Exon sequences
Seq C1 exon
GCTGCTTCAAACCAAATGCGGAATAGCAACGTCTTCCTGAAATTCATGTTCAACGAGGCCCTAAATGCGAATGGTGGACATCTCTTTGTGACCCACACGGCAGCTGAATGGCTGTTCGAAAGTTTCTACGACGAATTTTTGCATTACGCCATGACCCTGAATAATCCATTGGTCCCAAAAATCGAGTCGGATCACTTTGCCTGGTTCCTCAACCGTAACGGATCGAAGGACTTCGAGGGTCCCTTCACTGTACACACCGGAGTGGGTGACATCAAGGAGATGGGTGAAATTAAGTATTGGAAGGGCCAGAATCACACGGGATGGTACGATGGAGAATGTGGACGTCTCAATGGCAGCACCACGGATCTTTTTGTGCCCGATGAGCCCAAGGAGAAGGCTTTGACCATCTTCATTCCGGATACTTGTCGGATCATAAATCTTGAGTTCACCGGACAGAGCGAAACAATTCAGGGCATCACGGGTTGGAAGTACGAGATAACGCCGAATACCTTCGACAATGGACAGATCAATGGTAACGCGAAGTGCTGGTGTCCTTTGGAGCGCCAGCCAGATAATTGTCCTGCAACCGGAGCCACTGACCTAGGACCGTGTGCCGATGGAGTTCCCATGTACCTTTCTGCGGATCATTTTATGTACGCGGACGAAAGTTATGGCAGCACGATAAGCGGCTACAATCCGAATTACGATCAAAATAACTTTTATATCATCATGGAGCGCAAGATGGGAGTGCCGTTGAAGGTCAATGCCAATGTGATGATTACCTTGTACATAGAGCCAGACAGCGTTATAGA
Seq A exon
GTAATTATTTACAATAATTGTTGCCTACCATATATATGACTTGATAAACACATTTTTTCCTAG
Seq C2 exon
CATACTGAAGGGCCTGCCCAGCTTCTATGCACCACTTTTTACCACTGCTAGTCGAGCGGAAATCGACGAAGCCCTCGCCTCAGAACTTAGG
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0002939:FBtr0081031:3
Average complexity
IR
Mappability confidence:
NA
Protein Impact
ORF disruption upon sequence inclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.004 A=NA C2=0.000
Domain overlap (PFAM):
C1:
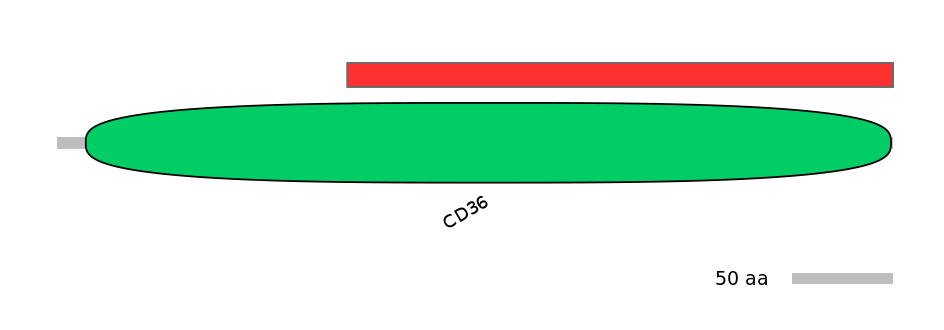
PF0113016=CD36=PD(67.3=99.3)
A:
NA
C2:
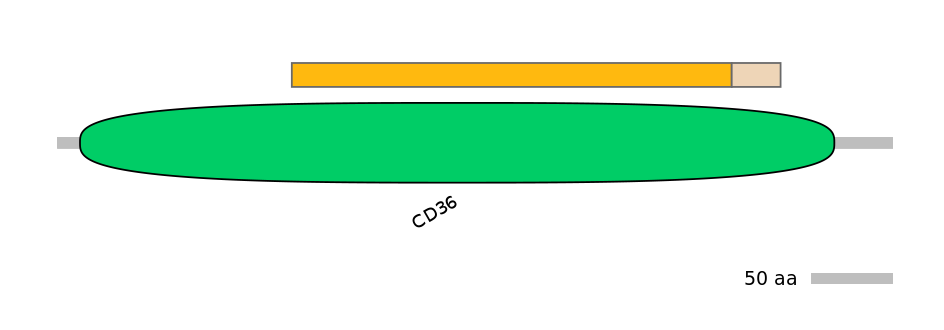
PF0113016=CD36=FE(6.5=100),PF149271=Neurensin=PU(11.7=22.6)
Main Inclusion Isoform:
FBpp0291642
Main Skipping Isoform:
FBpp0080584
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TCATCATGGAGCGCAAGATGG
R:
AGCAGTGGTAAAAAGTGGTGCA
Band lengths:
137-200
Functional annotations
There are 1 annotated functions for this event
PMID: 17087496
This event
This study show that the two splice variant isoforms encoded by ninaD exhibit different subcellular localizations. NinaD-I, the long protein variant, is localized at the plasma membrane, whereas the short variant, NinaD-II, is localized at intracellular membranes. Only NinaD-I could mediate the cellular uptake of carotenoids from micelles in this cell culture system. Carotenoid uptake was concentration-dependent and saturable. By in vivo analyses of different mutant and transgenic fly strains, the authors provide evidence of an essential role of NinaD-I in the absorption of dietary carotenoids to support visual chromophore synthesis. Moreover, our analyses suggest a role of NinaD-I in tocopherol metabolism. Even though Drosophila is a sterol auxotroph, the authors found no evidence of a contribution of NinaD-I to the uptake of these compounds. Note: annotated as APA, but DmeINT0034634 would have the same impact.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)