DreALTA0018783-1/2 @ danRer10
Alternative 3'ss
Gene
ENSDARG00000044532 | nr4a2b
Description
nuclear receptor subfamily 4, group A, member 2b [Source:ZFIN;Acc:ZDB-GENE-040718-103]
Coordinates
chr6:12227656-12228857:+
Coord C1 exon
chr6:12227656-12227754
Coord A exon
NA
Coord C2 exon
chr6:12228024-12228857
Length
0 bp
Sequences
Splice sites
5' ss Seq
GAGGTAAAC
5' ss Score
7.2
3' ss Seq
CAGCCATGCCCTGCGTCCAGGCT
3' ss Score
3.64
Exon sequences
Seq C1 exon
AAAAGCAAGCGAACGAAGAGACGCTCTGACATCAACCGGTACAAGAGCCCTGACTTCAGAGATTGTTCCACGAGAGATAAAAGTAGCAGGTGTGCGGAG
Seq A exon
NA
Seq C2 exon
GCTCAGTATGGGACATCACCTCCAGGAGCCAGCCCTGCATCTCAGAGTTACAGCTACAACACCACGGGAGAGTACAACTGCGACTTCTTAACGCCGGAGTTTGTCAAGTTTAGCATGGACCTGACTAACGCAGAGATAGCTGTCACTTCTTCGCTCCCCAGTTTCAGCACCTTCGTGGACACCTACAACTCCAACTACGACGTGAAGCCTCCCTGCCTCTACCAAATGGCCCACTCTGGGGATCAGCTGTCCATCAAGGTGGAGGAGATACCCGCACACGGCTACCATCAGCAGCAGCACCATCAGCCGCATCAGGCTGAGGAGAGCATACCCCATACCGGAGCTATATACTATAAACCATCCACTCCAAACATCTCCCAGTCCCCCAGTTACCCAACGGCTCCACATCATACGTGGGAAGACACCGGATCCCTTCACAGCTTTCACCAGAACTACCTGGCGACGTCACATATTATGGACCAGCAGCGGAAAAACGCGAT
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000044532-2-2,2-1-1/2
Average complexity
Alt3
Mappability confidence:
NA
Protein Impact
Protein isoform when splice site is used (No Ref, Alt. ATG)
No structure available
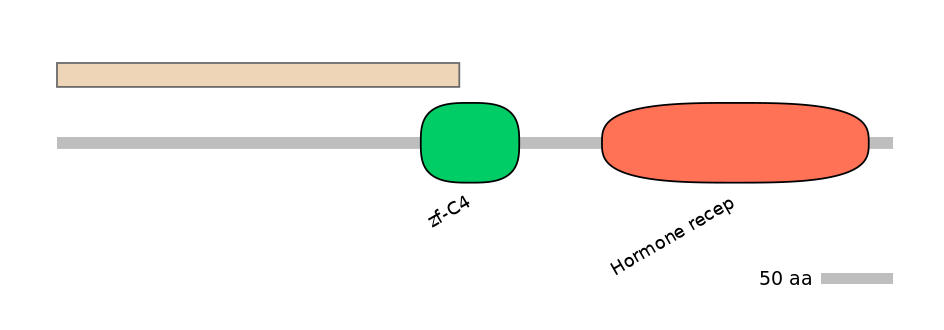
Features
Disorder rate (Iupred):
C1=NA A=NA C2=0.403
Domain overlap (PFAM):
C1:
NA
A:
NA
C2:
PF0010513=zf-C4=PU(38.6=9.5)
Main Inclusion Isoform:
NA
Other Inclusion Isoforms:
NA
Associated events
Conservation
Mouse
(mm9)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CGAACGAAGAGACGCTCTGAC
R:
CTCCTGGAGGTGATGTCCCAT
Band lengths:
118-135
Functional annotations
There are 1 annotated functions for this event
PMID: 16313515
Formed by alternative RNA splicing in exon 7 (HsaALTA1036233), nurr1a has a truncated carboxy-terminus, nurr1b has an internal deletion in the ligand-binding domain and nurr1c, newly identified in this study, has a novel carboxy-terminus produced by a frame shift downstream of the splice junction. Alternative RNA splicing in exon 3 (HsaALTA0005820) produces the isoform known as the transcriptionally-inducible nuclear receptor (TINUR), lacking the amino-terminus (when the internal Alt3 is used). Nurr2 and the newly identified nurr2c are produced by utilization of both exon 3 and exon 7 alternative splice sites. Transfection studies in dopaminergic SK-N-AS cells demonstrate that nurr1a, nurr1b, nurr1c and TINUR have significantly reduced transcriptional activities compared with full-length nurr1, while nurr2 and nurr2c are inactive. Furthermore, in these experiments, nurr2 and nurr2c both act as dominant negatives.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]