DreEX0026389 @ danRer10
Exon Skipping
Gene
ENSDARG00000068208 | def8
Description
differentially expressed in FDCP 8 homolog (mouse) [Source:ZFIN;Acc:ZDB-GENE-041114-96]
Coordinates
chr18:31469871-31481351:+
Coord C1 exon
chr18:31469871-31469980
Coord A exon
chr18:31471620-31471662
Coord C2 exon
chr18:31480841-31481351
Length
43 bp
Sequences
Splice sites
3' ss Seq
ATAATTTTTTTTGTTTTCAGCCG
3' ss Score
7.99
5' ss Seq
ATGGTAAAT
5' ss Score
6.41
Exon sequences
Seq C1 exon
AAATGCCAGGCCAAAGGATTCGTGTGTGAATTGTGTAAAGAGGGTGACAGTCTCTTTCCATTTGACAGCCACACCTCCGTTTGTCATGACTGCTTAGCAGTTTTCCACAG
Seq A exon
CCGCGCCACAGAGCGCCATCTGATTGCGCCATCTGGTTTCATG
Seq C2 exon
GGATTGTTACTATGATAACTCAACTATGTGTCCACATTGTGAGAGAATGCTGGAACACAAACTTGAGGTACCTTTGAAACTGTAGCAGAAGTGGGGCTGATCCCTGATGGATGTATGGCATACAAACACAGTGAAAGATCTTCTGAACGCATGTGGGTCTTGTCTCAGTTACTTTGTTTCTTCTCTTTATTTATTCTGAGATTTAAATCATGCATCTGTCAGCTATTGTGTGTCAGTGTTGATATTAATTCAGTTTCAGAAAAAAAAATACTAAAAAGCAAATTTATAGACATACTGAATATTGTCTATAAATTTATATATTAAAAACATAAATTGTGCTGCCAAATTATGTGGTCAGTGTGGTAAAGTTTTTATCTATTCATGTATCAGAAAAGGAATCTTCTGTAATGCAATTACTAATAACCAGCCATATACTATTAAAAGTCAGATTGCTGTTATTGCCTCTGTAATTACTGGAATTTAGTGAAACTGATGTCTTT
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000068208_MULTIEX1-10/10=9-C2
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
In the CDS, with uncertain impact
No structure available
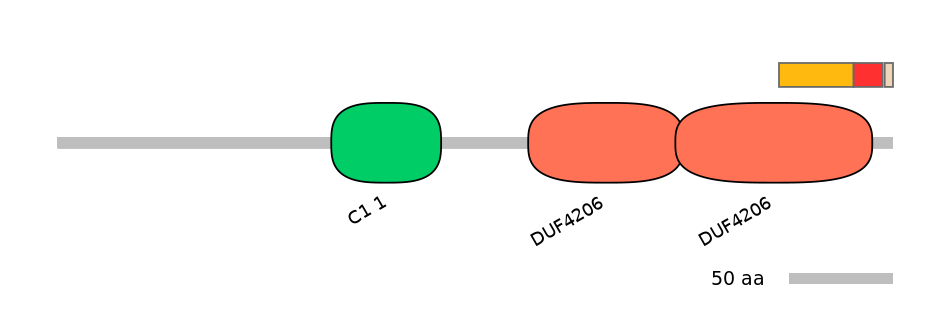
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
Domain overlap (PFAM):
C1:
PF139011=DUF4206=FE(33.6=100)
A:
PF139011=DUF4206=PD(9.4=60.0)
C2:
PF139011=DUF4206=PD(37.5=80.8)
Main Inclusion Isoform:
ENSDART00000170285fB2247
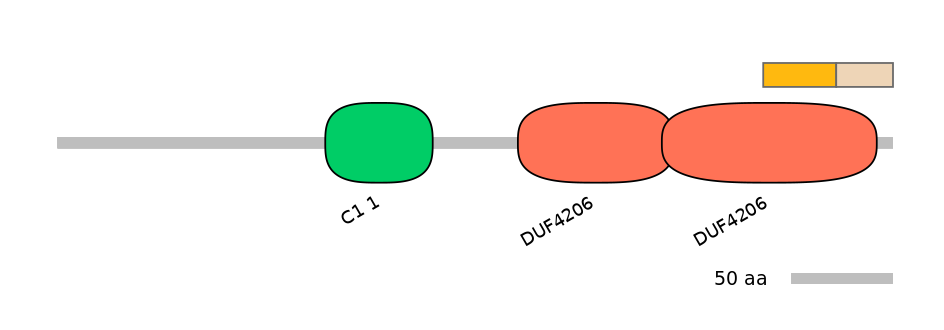
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ACCTCCGTTTGTCATGACTGC
R:
ATCAGCCCCACTTCTGCTACA
Band lengths:
139-182
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]