DreEX0027747 @ danRer10
Exon Skipping
Gene
ENSDARG00000010042 | dnm1a
Description
dynamin 1a [Source:ZFIN;Acc:ZDB-GENE-081104-27]
Coordinates
chr5:1267186-1290375:+
Coord C1 exon
chr5:1267186-1267253
Coord A exon
chr5:1275818-1275956
Coord C2 exon
chr5:1290289-1290375
Length
139 bp
Sequences
Splice sites
3' ss Seq
CCTCCATGATTCCTCTACAGGAC
3' ss Score
9.36
5' ss Seq
AAGGTAAAG
5' ss Score
9.06
Exon sequences
Seq C1 exon
ATGGAGTTTGATGAGAAGGAGCTGCGTAAAGAGATCAGCTACGCCATCAAGAACATCCACGGCATCAG
Seq A exon
GACGGGCCTGTTTACCCCTGACCTGGCCTTTGAGGCTATAGTGAAAAAGCAGATCCAAAAGCTAAACGGGCCGTGCCTGAAATGTATAGATATGGTTGTGAGCGAACTCACCTCCACCATTCGCAAATGTAGTGAGAAG
Seq C2 exon
CTGGCCCAGTATCCTATGCTGAGAGAAGAGATGGAGCGGATCGTCACACAGCACATCCGCGACCGCGAGAGCCGCACTAAAAATCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000010042_MULTIEX1-2/2=C1-C2
Average complexity
ME(1-2[100=100])
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.069
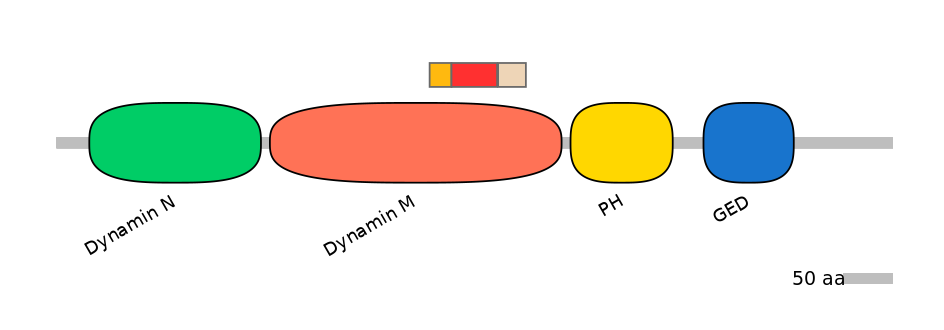
Domain overlap (PFAM):
C1:
PF0103115=Dynamin_M=FE(7.5=100)
A:
PF0103115=Dynamin_M=FE(15.6=100)
C2:
PF0103115=Dynamin_M=FE(9.5=100)
Other Skipping Isoforms:
NA
Associated events
Conservation
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TGGAGTTTGATGAGAAGGAGCTG
R:
CTGATTTTTAGTGCGGCTCTCG
Band lengths:
154-293
Functional annotations
There are 2 annotated functions for this event
PMID: 9725914
The authors generated stably transfected Clone 9 cells expressing full-length cDNAs of six different spliced forms of the three dynamin genes (Dnm1, Dnm2, and Dnm3) tagged with green fluorescent protein. The investigated splice variants are: Dnm1 (with mutually exclusive exon 10a (MmuEX6100535) or exon 10b (MmuEX0015279), Dnm2 (with/without cassette exon (MmuEX0015304), and Dnm3 (with/without cassette exon MmuEX0015311). Confocal or fluorescence microscopy of these transfected cells revealed that many of the dynamin proteins associate with distinct membrane compartments, which include clathrin-coated pits at the plasma membrane and the Golgi apparatus, and several undefined vesicle populations.
PMID: 20700442
[Negative results]. Dynamin-1 (Dnm1) encodes a large multimeric GTPase necessary for activity-dependent membrane recycling in neurons, including synaptic vesicle endocytosis. Mice heterozygous for a novel spontaneous Dnm1 mutation--fitful--experience recurrent seizures, and homozygotes have more debilitating, often lethal seizures in addition to severe ataxia and neurosensory deficits. Fitful is a missense mutation in an exon that defines the DNM1a isoform (uses Exon 10a (MmuEX6100535)), leaving intact the alternatively spliced exon that encodes DNM1b (uses exon 10b (MmuEX0015279). The expression of the corresponding alternate transcripts is developmentally regulated, with DNM1b expression highest during early neuronal development and DNM1a expression increasing postnatally with synaptic maturation. Mutant DNM1a does not efficiently self-assemble into higher order complexes known to be necessary for proper dynamin function, and it also interferes with endocytic recycling in cell culture. In mice, the mutation results in defective synaptic transmission characterized by a slower recovery from depression after trains of stimulation. The DNM1a and DNM1b isoform pair is highly conserved in vertebrate evolution, whereas invertebrates have only one isoform. The authors speculate that the emergence of more specialized forms of DNM1 may be important in organisms with complex neuronal function. The DNM1a and DNM1b isoforms show similar complex formation ability and localization.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]