DreEX0050000 @ danRer10
Exon Skipping
Gene
ENSDARG00000032630 | neb
Description
nebulin [Source:ZFIN;Acc:ZDB-GENE-041111-216]
Coordinates
chr9:23059700-23060254:-
Coord C1 exon
chr9:23060150-23060254
Coord A exon
chr9:23059916-23060023
Coord C2 exon
chr9:23059700-23059813
Length
108 bp
Sequences
Splice sites
3' ss Seq
CCTTTGTTGTATGTCCACAGACG
3' ss Score
10.19
5' ss Seq
GATGTAAGT
5' ss Score
9.11
Exon sequences
Seq C1 exon
CTCAACTACAAGGAAAAGTATGAGAAGGAGAAGTTTAAGTGTTACCTCCCTCCTGATTACCCTTTTTTCATTCAATCAAGAGTTAATGCGTATAACCTTAGTGAT
Seq A exon
ACGTGTTACAAGTATGACTGGGAAAAAACAAAAGCCAAGAAGTTTGAGGTGAAAGGAGATGCAATTTCGATCCGTGCAGCCCGCGCTCACACTAACATCGCTAGTGAT
Seq C2 exon
GTGAAATACAAGAAGGAATATGAGAAGAACAGAGGACAGATGGTCGGGGCCCTCAGTATCAAAGATGACCCCAAAATTATGCACTCAGTTCATGTTGCTAAGATCCAAAGCGAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000032630_CASSETTE2
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
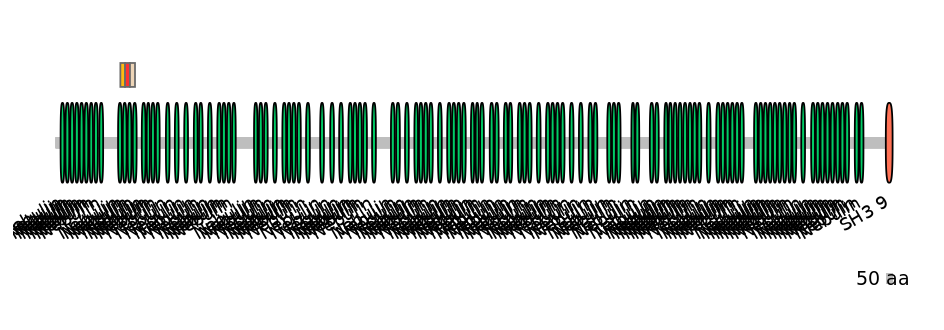
Domain overlap (PFAM):
C1:
PF0088013=Nebulin=PD(37.9=31.4),PF0088013=Nebulin=PU(55.2=45.7)
A:
PF0088013=Nebulin=PD(37.9=30.6),PF0088013=Nebulin=PU(55.2=44.4)
C2:
PF0088013=Nebulin=PD(37.9=28.9),PF0088013=Nebulin=PU(55.2=42.1)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TCAACTACAAGGAAAAGTATGAGAAG
R:
CTCGCTTTGGATCTTAGCAACA
Band lengths:
218-326
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]