DreEX0051772 @ danRer10
Exon Skipping
Gene
ENSDARG00000101234 | nsmfb
Description
NMDA receptor synaptonuclear signaling and neuronal migration factor b [Source:ZFIN;Acc:ZDB-GENE-080603-4]
Coordinates
chr5:27844928-27859132:+
Coord C1 exon
chr5:27844928-27845000
Coord A exon
chr5:27846300-27846305
Coord C2 exon
chr5:27859080-27859132
Length
6 bp
Sequences
Splice sites
3' ss Seq
ATCTTTCCTTGGGTCTAAAGAAT
3' ss Score
3.38
5' ss Seq
ATCGTGAGT
5' ss Score
8.88
Exon sequences
Seq C1 exon
AGGATCACACGAACAGCTGGTTTCCTAAAGAGAACATGTTCAGCTTCCAGACTGCGACCACCACCATGCAAGC
Seq A exon
AATATC
Seq C2 exon
AAACTTCCGGAAACATCTGAGGATGGTGGGCAGCAGGAGGGTGAAGGCCCAGA
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000101234-MICROEX1-1/2
Average complexity
MIC-M
Mappability confidence:
NA
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.702 A=0.333 C2=0.171
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
NO
Main Inclusion Isoform:
ENSDART00000165292fB4346
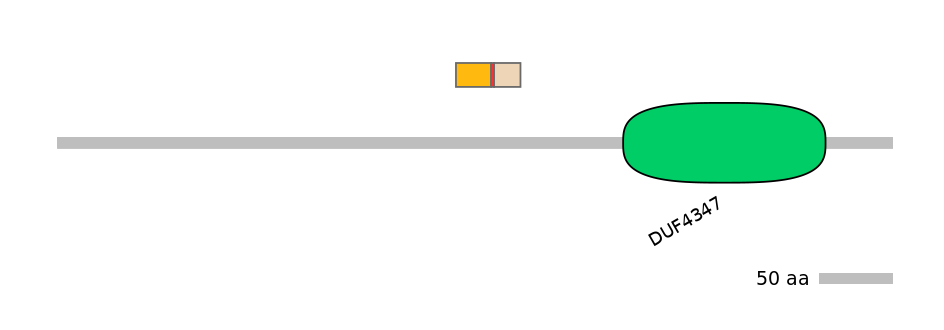
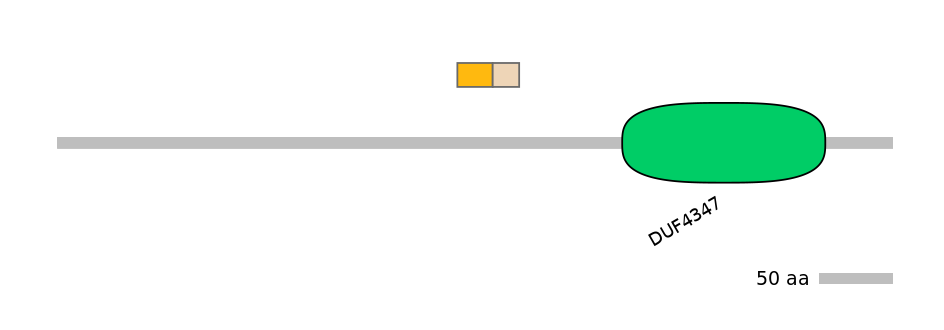
Other Inclusion Isoforms:
NA
Associated events
Conservation
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TCACACGAACAGCTGGTTTCC
R:
GCTGCCCACCATCCTCAGAT
Band lengths:
103-109
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]