DreEX0062945 @ danRer10
Exon Skipping
Gene
ENSDARG00000087962 | rpap3
Description
RNA polymerase II associated protein 3 [Source:ZFIN;Acc:ZDB-GENE-040426-928]
Coordinates
chr4:962486-965470:+
Coord C1 exon
chr4:962486-962556
Coord A exon
chr4:964112-964198
Coord C2 exon
chr4:965340-965470
Length
87 bp
Sequences
Splice sites
3' ss Seq
TGTGTGTGTTTTTATATCAGGAG
3' ss Score
8.48
5' ss Seq
ACCGTGAGT
5' ss Score
9.4
Exon sequences
Seq C1 exon
ATTTTGAGCAGGTTCTGAAGCTCGAGCCGGGAAACAAACAAGCCATAAGTGAGATCGAGAAGCTCACAGCG
Seq A exon
GAGATGCGCAGCAGTCTGGCTCCACAGGAGGACACACTGAGAAAGACCGTCCAGCCCATACACAAGCCTGAGCACCTCAGATCCACC
Seq C2 exon
AAACCCTTGAGGCGAATGGAGATTGAAGAGGTTGGAGGAGCGATCGCTCAGACTCCGCTCACGTCTTCACCTCATCATAAGATCCAGAAGATGGAGGAGAAGCCAGACGCCAGCTGTCCTGCTGTGGCCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000087962_MULTIEX1-1/2=C1-2
Average complexity
S*
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.708 A=1.000 C2=1.000
Domain overlap (PFAM):
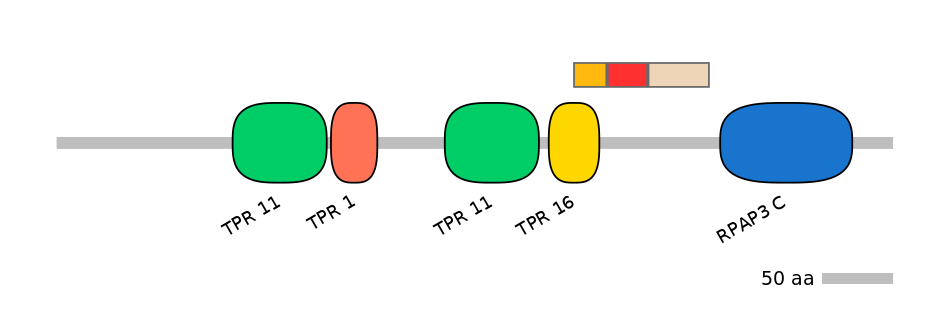
C1:
PF134321=TPR_16=PD(48.6=75.0)
A:
NO
C2:
NO
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GAGCAGGTTCTGAAGCTCGAG
R:
CGTCTGGCTTCTCCTCCATCT
Band lengths:
175-262
Functional annotations
There are 1 annotated functions for this event
PMID: 23159623
The exon is required for the protein's interaction with PIH1D1, which protects the cell against apoptosis. Knockdown of the isoform containing the exon down-regulates PIH1D1 protein level, without affecting mRNA level, suggesting the inclusion isoform stabilizes the R2AP3 protein complex (involved in cell survival) of which they both belong, whereas the skipping isoform has a dominant negative effect on the R2TP complex.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]