DreEX0073049 @ danRer10
Exon Skipping
Gene
ENSDARG00000028173 | slc4a2a
Description
solute carrier family 4 (anion exchanger), member 2a [Source:ZFIN;Acc:ZDB-GENE-051101-2]
Coordinates
chr2:32434158-32438749:-
Coord C1 exon
chr2:32438610-32438749
Coord A exon
chr2:32434875-32434931
Coord C2 exon
chr2:32434158-32434338
Length
57 bp
Sequences
Splice sites
3' ss Seq
AGCCTGCTGTTGTTTATTAGAGG
3' ss Score
4.9
5' ss Seq
AAGGTAGGG
5' ss Score
8.76
Exon sequences
Seq C1 exon
GTCACAGATTTGAAGATGTCCCAGGTGTGCGGCGGCACCTGGTTAAGAAGAGCGCTAAAGGACAGGTGGTTCACATCGGCAAGGACTACAAAGAGTTCAGCAGTCGTATTCGGACCAAGCTGGATCGAACTCCACATGAG
Seq A exon
AGGCGATCATGGCCCTGTTACATGGACCACAGGGTTCATCGTGCCCAGCAGCTGAAG
Seq C2 exon
GTTTTTGTTGAGCTGAATGAATTACTGATGGATAAGAACCAGGAGATGCATTGGAAAGAAACTGCTCGTTGGATAAAGTTTGAAGAGGATGTTGAGGAGGAGACTGAACGCTGGGGGAAACCTCATGTGGCTTCACTCTCGTTCCGCAGCCTGCTGGAGCTCCGCAAGACCATTTCACATG
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000028173_CASSETTE2
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.138 A=0.211 C2=0.000
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
PF075658=Band_3_cyto=PU(11.3=50.8)
Main Inclusion Isoform:
ENSDART00000041319fB6089
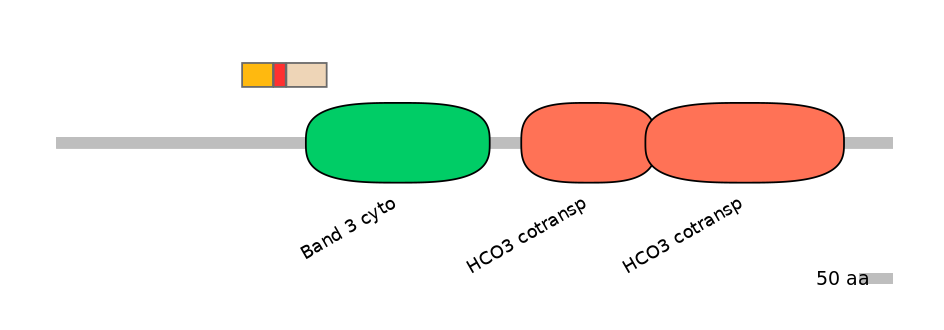
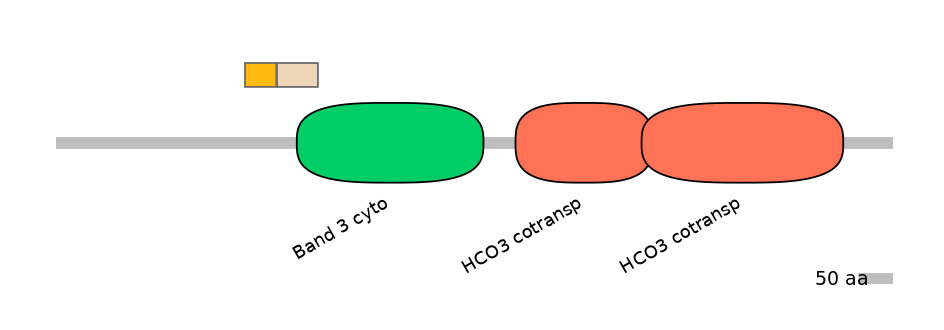
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CCAAGCTGGATCGAACTCCAC
R:
AGCGTTCAGTCTCCTCCTCAA
Band lengths:
138-195
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]