GgaALTA1026161-2/2 @ galGal4
Alternative 3'ss
Gene
ENSGALG00000006867 | QRICH1
Description
glutamine-rich 1 [Source:HGNC Symbol;Acc:HGNC:24713]
Coordinates
chr12:11599381-11600400:+
Coord C1 exon
chr12:11599381-11600157
Coord A exon
chr12:11600231-11600235
Coord C2 exon
chr12:11600236-11600400
Length
5 bp
Sequences
Splice sites
5' ss Seq
ATGGTGGGA
5' ss Score
2.88
3' ss Seq
TGCTAATTCACTCTTTCCAGCGC
3' ss Score
7.08
Exon sequences
Seq C1 exon
TCCAAGCACAGCTTGTGGCAGGACAGGCCATTACTGGTGGTCAGCAGATCCAGATCCAGACAGTGGGGGCGTTGTCACCTCCACCTTCTCAACAAGGCTCTCCGCGAGAGGGAGAGAGGAGGATCAGCACCGCTAGCGTCCTTCAGCCGGTGAAGAAACGCAAGGTGGATATGCCTATTACTGTATCGTATGCTATTTCAGGGCAGCCAGTTGCCACAGTGTTGGCCATTCCTCAGGGCCAGCAGCAAAGTTATGTCTCTCTAAGGCCAGACTTACTAACAGTGGACAGTGCCCACCTGTACAGTGCCACTGGGACCATTACTAGCCCCACTGGAGAGACTTGGACCATCCCTGTTTACTCTGCTCAGCCACGTGGTGACCTTCAGCAGCAAAACATAACCCACATCGCCATCCCTCAGGAGGCCTACAATGCTGTCCACGTCAGTGGCTCCCCAACGACACTGACCACAGTGAAGCTGGAAGATGACAAGGATAAGATG
Seq A exon
CGCAG
Seq C2 exon
TTTATGAATGGCAACATCCACATTCCAGTGGCAGTGCAGGCTGTGGCCGGGACATACCAGAACACAGCACAGACAGTGCACATATGGGACCCGCAGCAGCAACAGCAGCAGCCCACACCCCAAGAGCAGGGGCAGCAGCAGCAGCAACTACAAGTCACTTGCTCA
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000006867-14-10,14-9-2/2
Average complexity
Alt3
Mappability confidence:
NA
Protein Impact
ORF disruption when splice site is used (sequence inclusion)
No structure available
Features
Disorder rate (Iupred):
C1=0.270 A=NA C2=0.545
Domain overlap (PFAM):
C1:
NO
A:
NA
C2:
NO
Main Inclusion Isoform:
NA
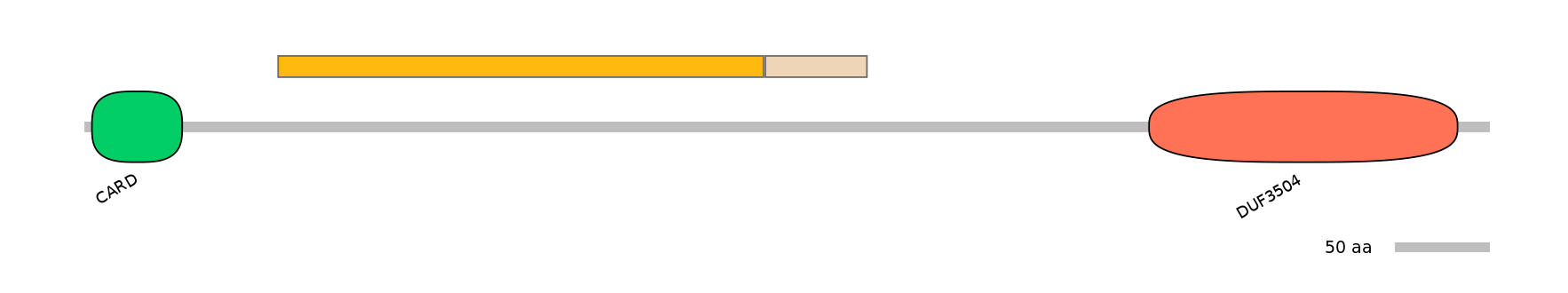
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GCCTACAATGCTGTCCACGTC
R:
ACTGGAATGTGGATGTTGCCA
Band lengths:
107-112
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]