GgaEX0009609 @ galGal3
Exon Skipping
Gene
ENSGALG00000006725 | ACAN
Description
NA
Coordinates
chr10:14737260-14747985:-
Coord C1 exon
chr10:14747195-14747985
Coord A exon
chr10:14739548-14739661
Coord C2 exon
chr10:14737260-14737418
Length
114 bp
Sequences
Splice sites
3' ss Seq
AAGCCCTTGATTTCTTGCAGACA
3' ss Score
6.49
5' ss Seq
TCGGTACGG
5' ss Score
8.77
Exon sequences
Seq C1 exon
AGAAGTAAGTGGGGAAAGTTCTGCCTTCCCTGAAATTAGCGTAGAGACATCCACAAGTCAGGAAGCCCGCGGTGAAACATCTGCCTTTCCTGAGATTGGCATAGAGACATCCACAGCCCACGAAGGCAGTGGGGAAACTCCTGGGCTGCCTGCTGTTAGCACTGACACTGCTGCCACGTCTCTGGCCAGTGGTGAGCCCTCCGGTGCTCCTGAGAAGGAAACTCCCGACACAACATCACATCTGATCACGGGCGTTTCAGGGGAAACCTCTGTCCCAGATGCTGTAATCAGTACCAGTGCTCCAGATGTTGAACTAGCGCAGGGACCCAGAAACACTGAAGAGACTCAGCTTGAAATAGAGCCCTCCACTCCTGCGGCATCTGGACAAGAGACAGAAACAGCTGCTGTCCTCGACAATCCCCATCTGCCAGCCACTGCTACTGCTGCCCTGCATCCAGCCTCCCAAGAAGCAGTAGATGCACTGGGACCCACGACAGAAG
Seq A exon
ACACTGATGAGTGCCACTCAAGCCCCTGTCTGAATGGAGCTACCTGCGTTGATGGCATCGACTCTTTCAAATGCTTATGCCTTCCCAGCTACGGAGGGGACCTGTGTGAGATCG
Seq C2 exon
ACCTGGCAAACTGTGAGGAAGGCTGGATCAAGTTCCAGGGCCACTGCTACAGGCACTTTGAAGAGAGGGAGACGTGGATGGACGCAGAGTCCAGGTGCAGAGAACATCAAGCCCACCTGAGCAGCATCATTACTCCAGAGGAACAAGAATTTGTGAACA
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000006725_CASSETTE1
Average complexity
S
Mappability confidence:
NA
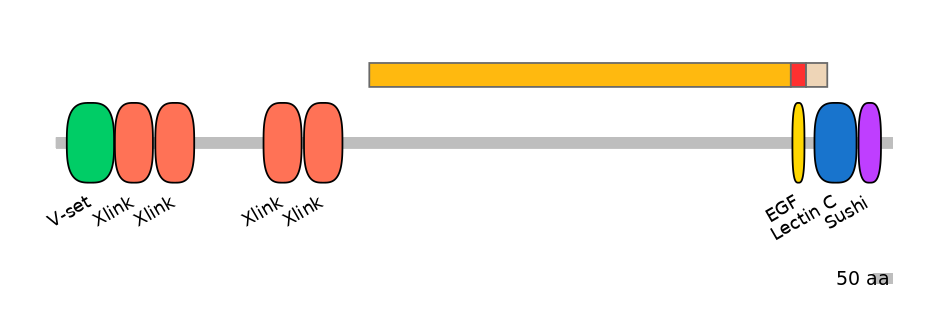
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.942 A=0.154 C2=0.000
Domain overlap (PFAM):
C1:
NO
A:
PF0000822=EGF=WD(100=79.5)
C2:
PF0005916=Lectin_C=PU(30.2=59.3)
Main Skipping Isoform:
NA
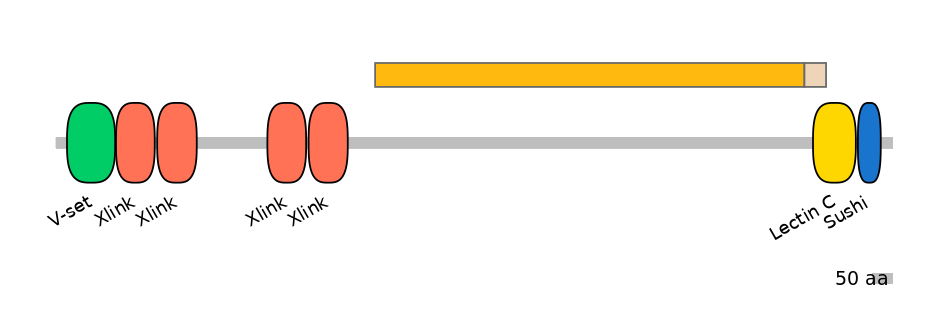
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
AACCTCTGTCCCAGATGCTGT
R:
CCTTCCTCACAGTTTGCCAGG
Band lengths:
258-372
Functional annotations
There are 1 annotated functions for this event
PMID: 14722076
The proteoglycans aggrecan, versican, neurocan, and brevican bind hyaluronan through their N-terminal G1 domains, and other extracellular matrix proteins through the C-type lectin repeat in their C-terminal G3 domains. Here the authors identify tenascin-C as a ligand for the lectins of all these proteoglycans and map the binding site on the tenascin molecule to fibronectin type III repeats, which corresponds to the proteoglycan lectin-binding site on tenascin-R. In the G3 domain, the C-type lectin is flanked by two epidermal growth factor (EGF) repeats and a complement regulatory protein-like motif. In aggrecan, the EGF repeats are subject to alternative splicing (each repeat is encoded by a cassette exon: EGF-like 1 is encoded by HsaEX0001787, and EGF-like 2 is encoded by HsaEX0001788). To investigate if these flanking modules affect the C-type lectin ligand interactions, the authors produced recombinant proteins corresponding to aggrecan G3 splice variants. The G3 variant proteins containing the C-type lectin showed different affinities for various ligands, including tenascin-C, tenascin-R, fibulin-1, and fibulin-2. The presence of an EGF motif enhanced the affinity of interaction, and in particular the splice variant containing both EGF motifs had significantly higher affinity for ligands, such as tenascin-R and fibulin-2.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]