GgaEX0026898 @ galGal3
Exon Skipping
Gene
ENSGALG00000023854 | Q90996_CHICK
Description
NA
Coordinates
chr17:3050578-3055747:-
Coord C1 exon
chr17:3055474-3055747
Coord A exon
chr17:3052990-3053265
Coord C2 exon
chr17:3050578-3050850
Length
276 bp
Sequences
Splice sites
3' ss Seq
TGTGTTCCTCTCTCCTTCAGAGG
3' ss Score
11.47
5' ss Seq
CAGGTACCT
5' ss Score
8.16
Exon sequences
Seq C1 exon
GTAGAGGAACCTCTCCTTAGCAAACTCACTGTATCAAATGCTACCTCAGACTCCATGTTGCTCATGTGGGAAGCACAGGACAATGCCTTTGACCATTTTATTCTAGAAGTCAGAAACTCTGACTCCCCTTTGGACTCCCTGGTGCAGATAGTGCCAGGGGCCTCCCGGCACTATGTAGTCACCAACCTCAAAGCTGCCACTAATTACACTGTCCAGCTCCATGGTGTAGTTGATGGGCAAGGTGGTCAGACCTTGACAGCACTGGCAACCACAG
Seq A exon
AGGCTGAGCCACAGCTGGGCACGCTCACACTCACAAATGTCACCCCCGACAGCTTCAACCTCTCGTGGACCACAAGAGATGGGCCTTTTGCCAAGTTTGTCATACACGTTAGAGATTCCTTTGCAGCACACGAACCCCAGGAGCTTACGGTGTCGGGAGGTGCTCGCAGCGCTCACATCTCAGGCCTGCTAGATTACACTGGCTATGACATTAACATTAAGGGCACCACCGATGCAGGTGTTCACACAGAGCCCCTCACTGCTTTTGTCATGACAG
Seq C2 exon
AGGCCATGCCCCCACTGGAAAACCTAACTGTCTCTGATATTAACCCTTACGGGTTCACAGTGTCATGGATGGCATCGGAGAATGCCTTCGACAGCTTTCTTGTAGTCGTGGTGGACTCTGGCAAGCTGCTGGACCCACAGGAGTTCCTCCTTACGGGAGCCCAGCGGCAACTGAAGCTTAAAGGCCTTATAACTGGCATTGGCTATGAGGTTATGCTTTATGGTTTTGCCAAGGGGCACCAAACAAAGCCCCTAAGCACTGTGGCTGTTACAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000023854-'0-1,'0-0,1-1
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
Show structural model
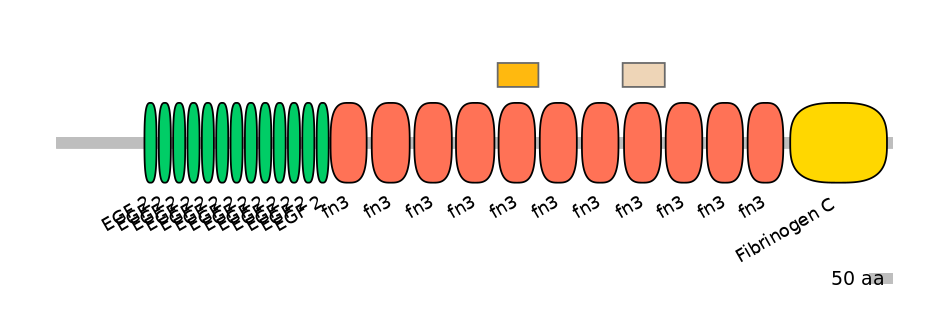
Features
Disorder rate (Iupred):
C1=0.087 A=0.075 C2=0.000
Domain overlap (PFAM):
C1:
PF0004116=fn3=WD(100=88.0)
A:
PF0004116=fn3=WD(100=88.2)
C2:
PF0004116=fn3=WD(100=91.2)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Mouse
(mm10)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CTCATGTGGGAAGCACAGGAC
R:
GCATTCTCCGATGCCATCCAT
Band lengths:
299-575
Functional annotations
There are 3 annotated functions for this event
PMID: 1725601
Three consecutive exons encoding one FN3 domain each. Modulation of binding to Fibronectin.
PMID: 9348542
This study examined the in vivo expression and cell adhesive properties of two full-length recombinant tenascin-C proteins: TN-190, which contains the eight constant fibronectin type III repeats, and TN-ADC, which contains the additional AD2 (HsaEX006150), AD1, and C (HsaEX0066151) repeats. In situ hybridization with probes specific for the AD2, AD1, and C repeats shows that these splice variants are expressed at sites of active tissue modeling and fibronectin expression in the developing avian feather bud and sternum. Transcripts incorporating the AD2, AD1, and C repeats are present in embryonic day 10 wing bud but not in embryonic day 10 lung. By using a panel of nine cell lines in attachment assays, the authors have found that C2C12, G8, and S27 myoblastic cells undergo concentration-dependent adhesion to both variants, organize actin microspikes that contain the actin-bundling protein fascin, and do not assemble focal contacts. On a molar basis, TN-ADC is more active than TN-190 in promoting cell attachment and irregular cell spreading. The addition of either TN-190 or TN-ADC in solution to C2C12, COS-7, or MG-63 cells adherent on fibronectin decreases cell attachment and results in decreased organization of actin microfilament bundles, with formation of cortical membrane ruffles and retention of residual points of substratum contact that contain filamentous actin and fascin. These data establish a biochemical similarity in the processes of cell adhesion to tenascin-C and thrombospondin-1, also an ?antiadhesive? matrix component, and also demonstrate that both the adhesive and adhesion-modulating properties of tenascin-C involve similar biochemical events in the cortical cytoskeleton. In addition to these generic properties, TN-ADC is less active in adhesion modulation than TN-190.
PMID: 11714809
Together, these 6-8 alternative exon array the encode minimal region of tenascin-C that can inhibit T cell activation. Recombinant fragments corresponding to defined regions of the molecule were tested for their ability to inhibit in vitro activation of human peripheral blood T cells induced by anti-CD3 mAbs in combination with fibronectin or IL-2. A recombinant protein encompassing the alternatively spliced fibronectin type III domains of tenascin-C (TnFnIII A1-3 and B-D) vigorously inhibited both early and late lymphocyte activation events including activation-induced TCR/CD8 down-modulation, cytokine production, and DNA synthesis. In agreement with this, full length recombinant tenascin-C containing the alternatively spliced region suppressed T cell activation, whereas tenascin-C lacking this region did not. Using a series of smaller fragments and deletion mutants issued from this region, the authors have identified the TnFnIII A1A2 domain as the minimal region suppressing T cell activation. Single TnFnIII A1 or A2 domains were no longer inhibitory, while maximal inhibition required the presence of the TnFnIII A3 domain.