GgaEX1012824 @ galGal4
Exon Skipping
Gene
ENSGALG00000009382 | CAPN8
Description
calpain 8 [Source:HGNC Symbol;Acc:HGNC:1485]
Coordinates
chr3:17058883-17060409:+
Coord C1 exon
chr3:17058883-17058968
Coord A exon
chr3:17059268-17059392
Coord C2 exon
chr3:17060335-17060409
Length
125 bp
Sequences
Splice sites
3' ss Seq
CTCAACATAACTTTTCTCAGAAG
3' ss Score
2.92
5' ss Seq
CTGGTCAGT
5' ss Score
6.6
Exon sequences
Seq C1 exon
GTCTATTACCGAGGACGGCCAGAGAAACTTGTGAGGCTTAGAAATCCCTGGGGTGAAATAGAATGGACTGGAGCGTGGAGTGATAA
Seq A exon
AAGATCCCATTGGTGCTACCAAGACCTTAGCAGGACAGGCAAACCAATTCCATTACTGGAAAACACAGCAAGGCTACACTGCTGTGCCCCTAAGTCAGCTTTGAAGTTTTGGAGTTCTAAATCTG
Seq C2 exon
TGCTCCCGAATGGAATTATATTGATCCCAAACAAAAGCAGGCTCTGGATAAGCAAGTAGATGATGGAGAATTTTG
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000009382_CASSETTE2
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
In the CDS, with uncertain impact
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.068
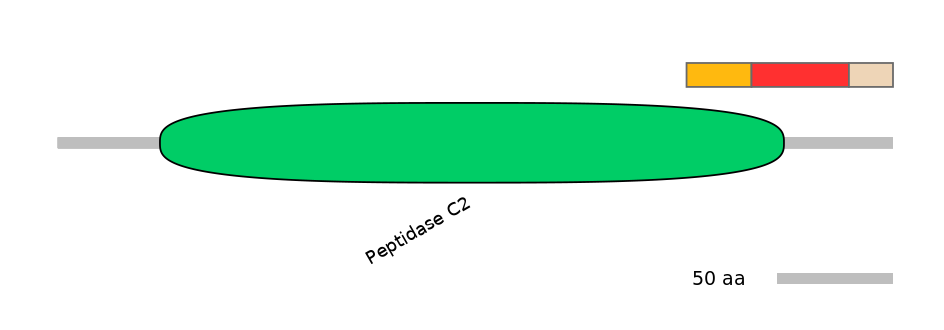
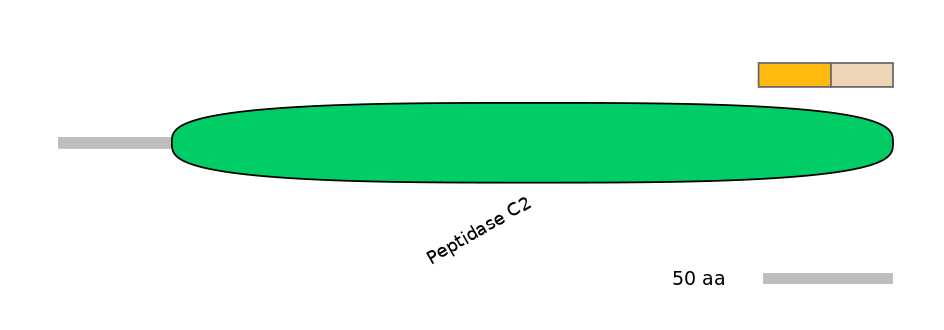
Domain overlap (PFAM):
C1:
PF0064816=Peptidase_C2=FE(10.0=100)
A:
PF0064816=Peptidase_C2=PD(5.2=32.6)
C2:
PF0064816=Peptidase_C2=FE(8.6=100)
Main Inclusion Isoform:
ENSGALT00000015288fB5745
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GAGGACGGCCAGAGAAACTTG
R:
TTCTCCATCATCTACTTGCTTATCCA
Band lengths:
146-271
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]