GgaEX1031316 @ galGal3
Exon Skipping
Gene
ENSGALG00000012537 | F1NCD3_CHICK
Description
NA
Coordinates
chr7:37363230-37392741:+
Coord C1 exon
chr7:37363230-37363923
Coord A exon
chr7:37366915-37367131
Coord C2 exon
chr7:37392151-37392741
Length
217 bp
Sequences
Splice sites
3' ss Seq
CTCCCTCTGCTCTCCCCCAGTCC
3' ss Score
10.53
5' ss Seq
CGGGTGAGT
5' ss Score
9.89
Exon sequences
Seq C1 exon
TGTCCGACCTCTTCACCACGCTGGTGGACCTCAAGTGGCGCTGGAACCTCTTCATTTTCGTCCTCACCTACACCGTGGCCTGGCTCTTCATGGCCTCCATGTGGTGGGTGATCGCCTACATGCGGGGCGACCTGAACAAGGCTCACGACGACAGCTACACCCCCTGCGTGGCCAACGTCTACAACTTCCCTTCCGCCTTCCTCTTCTTCATCGAGACCGAGGCCACCATCGGCTACGGCTACCGCTACATCACGGACAAATGCCCCGAGGGCATCATCCTCTTCCTCTTCCAGTCCATCCTGGGCTCCATCGTGGACGCCTTCCTCATTGGCTGCATGTTCATCAAGATGTCCCAGCCCAAGAAGAGGGCTGAGACGCTGATGTTCAGCGAGCACGCGGCCATCTCCATGCGGGACGGCAAGCTCACCCTCATGTTCCGCGTGGGGAACCTCCGCAACAGCCATATGGTCTCAGCGCAGATCCGCTGCAAGCTGCTCAAG
Seq A exon
TCCCGCCAGACGCCCGAGGGTGAGTTCCTGCCGCTGGACCAGCTGGAGCTGGACGTGGGCTTCAGCACAGGTGCCGACCAGCTCTTCCTCGTCTCGCCGCTCACCATCTGCCACGTCATCGATGCCAAGAGCCCCTTCTACGACCTGTCCCAGCGCAGCATGCAGACGGAGCAGTTCGAGATCGTCGTCATCCTGGAGGGCATCGTGGAAACCACGG
Seq C2 exon
GGATGACGTGCCAGGCCAGGACATCCTACACTGAGGATGAGGTGCTCTGGGGCCATCGCTTCTTCCCTGTTATTTCCTTGGAAGAAGGGTTCTTCAAAGTCGACTACTCGCAGTTCCACGCGACGTTTGAGGTCCCCACGCCGCCGTACAGCGTGAAGGAGCAGGAGGAGATGCTGCTCATGTCCTCACCCCTGATAGCGCCCGCTGTCAGCAACAGCAAGGAGAGGAATAACTCGGTGGAGTGCCTGGATGGTCTGGATGAGGTTGGTATAAAACTCCCTTCCAAACTGCAGAAAATAACTGGAAGGGACGACTTCCCCAAAAAACTCCTCAGGATGAGCTCCACCACCTCGGAGAAGGCCTACAGCATGGGTGATTTGCCCATGAAGCTGCAGCGGATCAGCTCAGTCCCTGGGAATTCAGAAGAGAAACTGGTGTCCAAAGCCACCAAGATGATGTCGGATCCCATGAGCCAGTCGGTGGCCGACTTGCCCCCCAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000012537-'0-5,'0-1,1-5=AN
Average complexity
A_S
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
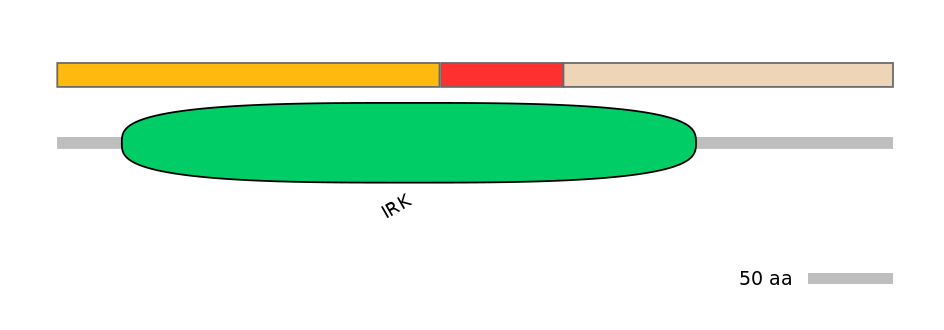
Features
Disorder rate (Iupred):
C1=0.058 A=0.000 C2=0.402
Domain overlap (PFAM):
C1:
PF0100715=IRK=PU(55.2=82.7),PF117553=DUF3311=WD(100=16.4)
A:
PF0100715=IRK=FE(21.2=100)
C2:
PF0100715=IRK=PD(23.0=40.0)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TACATCACGGACAAATGCCCC
R:
CCAGAGCACCTCATCCTCAGT
Band lengths:
305-522
Functional annotations
There are 3 annotated functions for this event
PMID: 18097938
This study examines two GIRK1 channel splice isoforms: GIRK1a (referred to as GIRK1), and GIRK1d. The GIRK1d variant lacks a region corresponding to exon 2 of full-length GIRK1 (encoded by HsaEX0033023), leading to a truncated GIRK1 that lacks the main part of the C-terminus. To study GIRK1d the authors used the Xenopus laevis expression system, the two-electrode voltage clamp method, and confocal laser scan microscopy. The authors found that these GIRK1d variant preferentially binds GIRK2 or GIRK4 over GIRK1. Furthermore, it largely reduces conductances mediated by GIRK1/2 or GIRK1/4 hetero-multimeric channels when coexpressed and nearly totally abolishes currents when replacing GIRK1 in hetero-multimeric channels.
PMID: 20512921
The aim of this study was to investigate the impact of increased mRNA levels encoding GIRK1 in breast tumours on GIRK protein expression. mRNA levels encoding hGIRK1 and hGIRK4 in the MCF7, MCF10A and MDA_MB_453 breast cancer cell lines were assessed and the corresponding proteins detected using Western blots. cDNAs encoding for four hGIRK1 splice variants (hGIRK1a (uses cassette exon HsaEX0033023), 1c, 1d (differs from 1a in skipping cassette exon HsaEX0033023) and 1e) were cloned from the MCF7 cell line. Subcellular localisation of fluorescence labelled hGIRK1a?e and hGIRK4 and of endogenous GIRK1 and GIRK4 subunits was monitored in the MCF7 cell line. All hGIRK1 splice variants and hGIRK4 were predominantly located within the endoplasmic reticulum. Heterologous expression in Xenopus laevis oocytes and two electrode voltage clamp experiments together with confocal microscopy were performed. Only the hGIRK1a subunit was able to form functional GIRK channels in connection with hGIRK4. The other splice variants are expressed, but exert a dominant negative effect on heterooligomeric channel function
PMID: 27519272
In order to survey possible tumorigenic properties of G-protein activated inwardly rectifying K+ channel (GIRK1) overexpression (encoded by the KCNJ3 gene), a range of malignant mammary epithelial cells, based on the MCF-7 cell line that permanently overexpress different splice variants of the KCNJ3 gene (GIRK1a (uses cassette exon HsaEX0033023), GIRK1c, GIRK1d (differs from 1a in skipping HsaEX0033023) and as a control, eYFP) were produced. Subsequently, selected cardinal neoplasia associated cellular parameters were assessed and compared: Adhesion to fibronectin coated surface as well as cell proliferation remained unaffected. Other vital parameters intimately linked to malignancy, i.e. wound healing, chemoinvasion, cellular velocities / motilities and angiogenesis were massively affected by GIRK1 overexpression. Overexpression of different GIRK1 splice variants exerted differential actions. While GIRK1a and GIRK1c overexpression reinforced the affected parameters towards malignancy, overexpression of GIRK1d resulted in the opposite. Single channel recording using the patch clamp technique revealed functional GIRK channels in the plasma membrane of MCF-7 cells albeit at very low frequency. The authors conclude that GIRK1d acts as a dominant negative constituent of functional GIRK complexes present in the plasma membrane of MCF-7 cells, while overexpression of GIRK1a and GIRK1c augmented their activity.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]