HsaALTA0005820-1/2 @ hg19
Alternative 3'ss
Gene
ENSG00000153234 | NR4A2
Description
nuclear receptor subfamily 4, group A, member 2 [Source:HGNC Symbol;Acc:7981]
Coordinates
chr2:157186426-157187303:-
Coord C1 exon
chr2:157187180-157187303
Coord A exon
NA
Coord C2 exon
chr2:157186426-157186522
Length
0 bp
Sequences
Splice sites
5' ss Seq
AAGGTTAGT
5' ss Score
8.54
3' ss Seq
GCCACCACTTCTCTCCCCAGCTT
3' ss Score
7.43
Exon sequences
Seq C1 exon
GGAGGAGATTGGACAGGCTGGACTCCCCATTGCTTTTCTAAAAATCTTGGAAACTTTGTCCTTCATTGAATTACGACACTGTCCACCTTTAATTTCCTCGAAAACGCCTGTAACTCGGCTGAAG
Seq A exon
NA
Seq C2 exon
CTTCAGTACCTTTATGGACAACTACAGCACAGGCTACGACGTCAAGCCACCTTGCTTGTACCAAATGCCCCTGTCCGGACAGCAGTCCTCCATTAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000153234-3-11,3-9-1/2
Average complexity
Alt3
Mappability confidence:
NA
Protein Impact
Protein isoform when splice site is used (No Ref, Alt. ATG)
No structure available
Features
Disorder rate (Iupred):
C1=NA A=NA C2=0.563
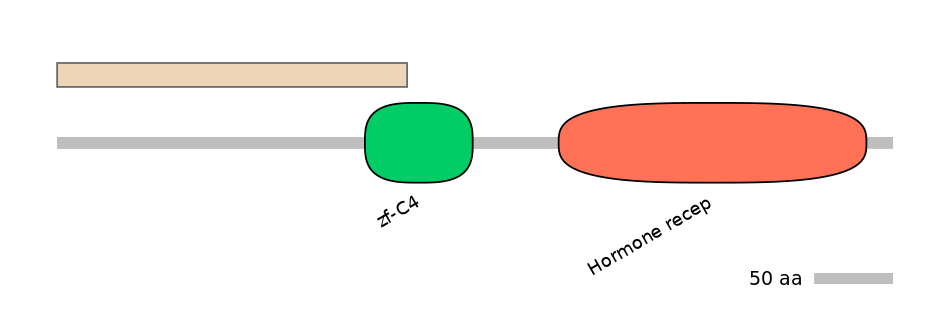
Domain overlap (PFAM):
C1:
NO
A:
NA
C2:
PF0010513=zf-C4=PU(38.6=12.0)
Other Skipping Isoforms:
Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GAGGAGATTGGACAGGCTGGA
R:
GTCCGGACAGGGGCATTTG
Band lengths:
203-381
Functional annotations
There are 2 annotated functions for this event
PMID: 16313515
Formed by alternative RNA splicing in exon 7 (HsaALTA1036233), nurr1a has a truncated carboxy-terminus, nurr1b has an internal deletion in the ligand-binding domain and nurr1c, newly identified in this study, has a novel carboxy-terminus produced by a frame shift downstream of the splice junction. Alternative RNA splicing in exon 3 (HsaALTA0005820) produces the isoform known as the transcriptionally-inducible nuclear receptor (TINUR), lacking the amino-terminus (when the internal Alt3 is used). Nurr2 and the newly identified nurr2c are produced by utilization of both exon 3 and exon 7 alternative splice sites. Transfection studies in dopaminergic SK-N-AS cells demonstrate that nurr1a, nurr1b, nurr1c and TINUR have significantly reduced transcriptional activities compared with full-length nurr1, while nurr2 and nurr2c are inactive. Furthermore, in these experiments, nurr2 and nurr2c both act as dominant negatives.
PMID: 16313515
Formed by alternative RNA splicing in exon 7 (HsaALTA1036233), nurr1a has a truncated carboxy-terminus, nurr1b has an internal deletion in the ligand-binding domain and nurr1c, newly identified in this study, has a novel carboxy-terminus produced by a frame shift downstream of the splice junction. Alternative RNA splicing in exon 3 (HsaALTA0005820) produces the isoform known as the transcriptionally-inducible nuclear receptor (TINUR), lacking the amino-terminus (when the internal Alt3 is used). Nurr2 and the newly identified nurr2c are produced by utilization of both exon 3 and exon 7 alternative splice sites. Transfection studies in dopaminergic SK-N-AS cells demonstrate that nurr1a, nurr1b, nurr1c and TINUR have significantly reduced transcriptional activities compared with full-length nurr1, while nurr2 and nurr2c are inactive. Furthermore, in these experiments, nurr2 and nurr2c both act as dominant negatives.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)