HsaALTD0004534-1/2 @ hg19
Alternative 5'ss
Gene
ENSG00000070756 | PABPC1
Description
poly(A) binding protein, cytoplasmic 1 [Source:HGNC Symbol;Acc:8554]
Coordinates
chr8:101730001-101730508:-
Coord C1 exon
chr8:101730459-101730508
Coord A exon
NA
Coord C2 exon
chr8:101730001-101730116
Length
0 bp
Sequences
Splice sites
5' ss Seq
CCAGTACGC
5' ss Score
3.88
3' ss Seq
GGGTATATGTTTTTCCCTAGGTG
3' ss Score
10.19
Exon sequences
Seq C1 exon
CGGAGCGTGCTTTGGACACCATGAATTTTGATGTTATAAAGGGCAAGCCA
Seq A exon
NA
Seq C2 exon
GTGGTTTGTGATGAAAATGGTTCCAAGGGCTATGGATTTGTACACTTTGAGACGCAGGAAGCAGCTGAAAGAGCTATTGAAAAAATGAATGGAATGCTCCTAAATGATCGCAAAGT
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000070756-10-12,11-12-1/2
Average complexity
Alt5
Mappability confidence:
NA
Protein Impact
Protein isoform when splice site is used (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.020 A=NA C2=0.002
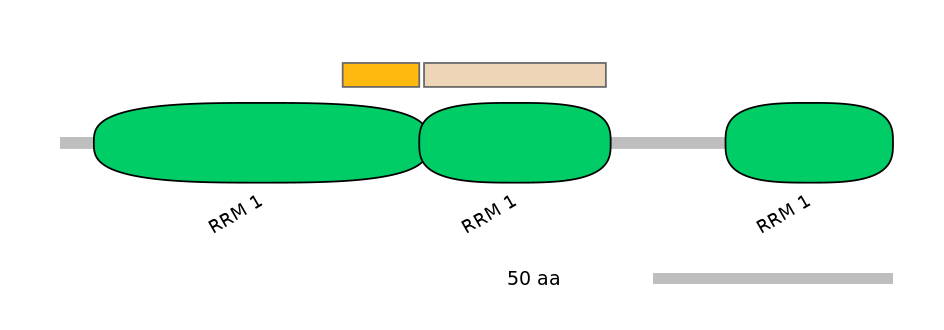
Domain overlap (PFAM):
C1:
PF0007617=RRM_1=FE(23.9=100),PF0007617=RRM_1=PU(0.1=0.0)
A:
NA
C2:
PF0007617=RRM_1=PD(2.8=5.1),PF0007617=RRM_1=PU(92.7=97.4)
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
Associated events
Conservation
Rat
(rn6)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TGATGTTATAAAGGGCAAGCCA
R:
TGCGATCATTTAGGAGCATTCCA
Band lengths:
134-182
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)