HsaEX0001787 @ hg19
Exon Skipping
Gene
ENSG00000157766 | ACAN
Description
aggrecan [Source:HGNC Symbol;Acc:319]
Coordinates
chr15:89399751-89414771:+
Coord C1 exon
chr15:89399751-89402648
Coord A exon
chr15:89403557-89403670
Coord C2 exon
chr15:89414613-89414771
Length
114 bp
Sequences
Splice sites
3' ss Seq
CTCTCCCCTGGGGGTTGCAGCCC
3' ss Score
4.93
5' ss Seq
TAGGTAAGG
5' ss Score
9.31
Exon sequences
Seq C1 exon
CGGGGCATCTGCCGCCCCTGAGGCCAGCAGAGAAGATTCTGGGTCCCCTGATCTGAGTGAAACCACCTCTGCATTCCACGAAGCTAACCTTGAGAGATCCTCTGGCCTAGGAGTGAGCGGCAGCACTTTGACATTTCAAGAAGGCGAGGCGTCCGCTGCCCCAGAAGTGAGTGGAGAATCCACCACCACCAGTGATGTGGGGACAGAGGCACCAGGCTTGCCTTCAGCCACTCCCACGGCTTCTGGAGACAGGACTGAAATCAGCGGAGACCTGTCTGGTCACACCTCGCAGCTGGGCGTTGTCATCAGCACCAGCATCCCAGAGTCTGAGTGGACCCAGCAGACCCAGCGCCCTGCAGAGACGCATCTAGAAATTGAGTCCTCAAGCCTCCTGTACTCAGGAGAAGAGACTCACACAGTCGAAACAGCCACCTCCCCAACAGATGCTTCCATCCCAGCTTCTCCGGAATGGAAACGTGAATCAGAATCAACTGCTGCAG
Seq A exon
CCCCCGCCAGGTCCTGTGCAGAGGAGCCCTGTGGAGCTGGGACCTGCAAGGAGACAGAGGGACACGTCATATGCCTGTGCCCCCCTGGCTACACTGGCGAGCACTGTAACATAG
Seq C2 exon
ACCAGGAGGTATGTGAGGAGGGCTGGAACAAGTACCAGGGCCACTGTTACCGCCACTTCCCGGACCGCGAGACCTGGGTGGATGCTGAGCGCCGGTGTCGGGAGCAGCAGTCACACCTGAGCAGCATCGTCACCCCCGAGGAGCAGGAGTTTGTCAACA
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000157766_MULTIEX1-1/2=C1-C2
Average complexity
C1
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.954 A=0.179 C2=0.244
Domain overlap (PFAM):
C1:
NO
A:
PF0000822=EGF=WD(100=78.9),PF0000822=EGF=PU(5.3=5.3)
C2:
PF0005916=Lectin_C=PU(30.2=59.3)

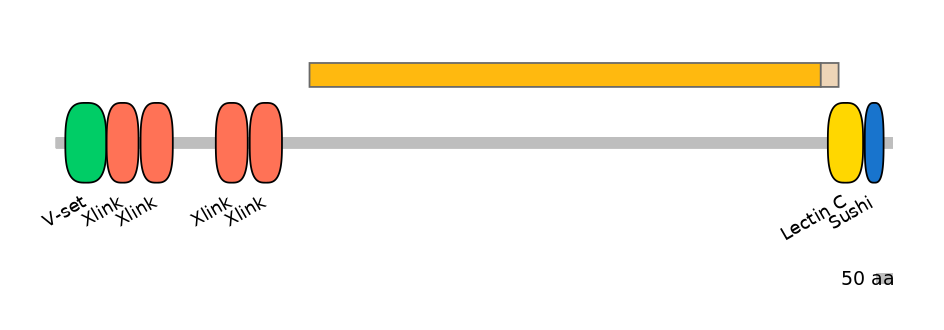
Associated events
Other assemblies
Conservation
Rat
(rn6)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TTGTCATCAGCACCAGCATCC
R:
CAGTGGCCCTGGTACTTGTTC
Band lengths:
247-361
Functional annotations
There are 1 annotated functions for this event
PMID: 14722076
This event
The proteoglycans aggrecan, versican, neurocan, and brevican bind hyaluronan through their N-terminal G1 domains, and other extracellular matrix proteins through the C-type lectin repeat in their C-terminal G3 domains. Here the authors identify tenascin-C as a ligand for the lectins of all these proteoglycans and map the binding site on the tenascin molecule to fibronectin type III repeats, which corresponds to the proteoglycan lectin-binding site on tenascin-R. In the G3 domain, the C-type lectin is flanked by two epidermal growth factor (EGF) repeats and a complement regulatory protein-like motif. In aggrecan, the EGF repeats are subject to alternative splicing (each repeat is encoded by a cassette exon: EGF-like 1 is encoded by HsaEX0001787, and EGF-like 2 is encoded by HsaEX0001788). To investigate if these flanking modules affect the C-type lectin ligand interactions, the authors produced recombinant proteins corresponding to aggrecan G3 splice variants. The G3 variant proteins containing the C-type lectin showed different affinities for various ligands, including tenascin-C, tenascin-R, fibulin-1, and fibulin-2. The presence of an EGF motif enhanced the affinity of interaction, and in particular the splice variant containing both EGF motifs had significantly higher affinity for ligands, such as tenascin-R and fibulin-2.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)