HsaEX0026914 @ hg19
Exon Skipping
Gene
ENSG00000144278 | GALNT13
Description
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 (GalNAc-T13) [Source:HGNC Symbol;Acc:23242]
Coordinates
chr2:155295104-155310361:+
Coord C1 exon
chr2:155295104-155295238
Coord A exon
chr2:155303080-155303277
Coord C2 exon
chr2:155306923-155310361
Length
198 bp
Sequences
Splice sites
3' ss Seq
TTATTTTCTTTGTCAAACAGACC
3' ss Score
8.69
5' ss Seq
GAGGTTTGT
5' ss Score
6.36
Exon sequences
Seq C1 exon
GTATTTTCTTACACTGCTGACAAAGAAATCCGAACCGATGACTTGTGCTTGGATGTTTCTAGACTCAATGGACCTGTAATCATGTTAAAATGCCACCATATGAGAGGAAATCAGTTATGGGAATATGATGCTGAG
Seq A exon
ACCCACACTCTTCTTCATATAATCACCCAGTCTTGTCTCTCAGTGAACAAAGTAGCTGATGGCTCCCAGCATCCTACTGTGGAAACCTGTAATGATAGCACTTTGCAAAAATGGCTACTAAGAAACTATACAAGAATGGAAATTTTTAGAAATATTTTTGGGAATTCTACTGATTACATTCTCTAATGATTTTTGGAG
Seq C2 exon
AGACTCACGTTGCGACATGTTAACAGTAACCAATGTCTCGATGAACCTTCTGAAGAAGACAAAATGGTGCCTACAATGCAGGACTGTAGTGGAAGCAGATCCCAACAGTGGCTGCTAAGGAACATGACCTTGGGCACATGAAGATCATGTCCTCCAAGCCATGAAAGTGTCTACGCTTTTGTTTTTCCATTATTTCAATTGGGGGAAAATATTAACTTTGCTGAATTGAAAGTTTTAAAAATCCTTTTAGTATTCTAAAACACAATTGTTTCTAATTCGTTTCTAGAAATGTTTGCTTATTTCCCTACTAAAATTTGTATCTGATCAAAGCACATAAGAATATAAATAATAGCAAACTACTATTAAACAACAGAACAACTTGTAAAACAAATTGTGTTTGCTTTAAGAAAAATGTTTATTGCACTCATGTCATAGGGTTAATTGGAGGTTATTTTATTTTTGGTTGTCATGGTGATTGAAAGAGATAATGTAAATGCCTT
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000144278_CASSETTE2
Average complexity
S*
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref, Alt. Stop)
No structure available
Features
Disorder rate (Iupred):
C1=0.007 A=0.010 C2=0.285
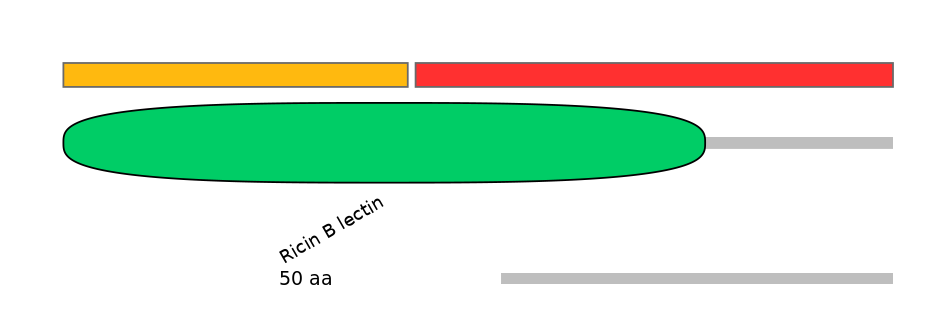
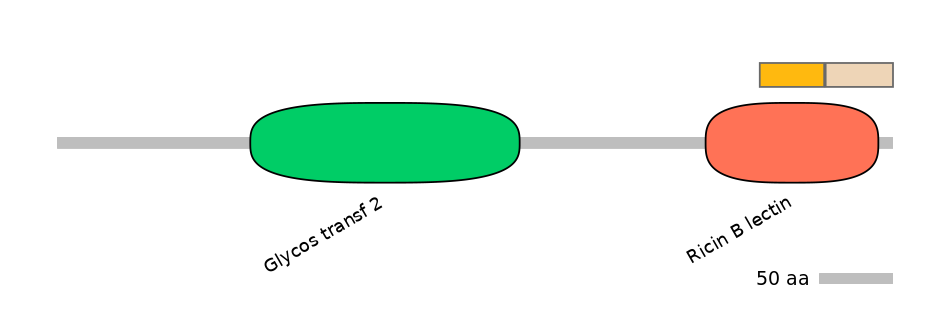
Domain overlap (PFAM):
C1:
PF0065217=Ricin_B_lectin=PU(53.0=97.8)
A:
PF0065217=Ricin_B_lectin=PD(27.8=61.5)
C2:
PF0065217=Ricin_B_lectin=PD(30.3=76.6)
Associated events
Other assemblies
Conservation
Rat
(rn6)
No conservation detected
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ATCCGAACCGATGACTTGTGC
R:
ATGTGCCCAAGGTCATGTTCC
Band lengths:
247-445
Functional annotations
There are 2 annotated functions for this event
PMID: 22186971
This event
The study identifies and characterizes enzymatically active splice variants of GalNAc-T13 that differ in the sequence of their lectin domain (via inclusion/exclusion a penultimate exon that changes C-terminus (HsaEX0026914)). The variants differ in their affinities for glycopeptide substrates.
PMID: 27913570
This event
Polypeptide GalNAc-transferase 13 (GalNAc-T13) splice variants with changes in their lectin domains were characterized by in vitro glycosylation assay: one (delta39Ex9) (alternative splice acceptor site in exon 9, not in VastDB) was inactive while the second one (Ex10b) (cassette exon, HsaEX0026914) had essentially unaltered activity.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)