HsaEX0032352 @ hg19
Exon Skipping
Gene
ENSG00000005884 | ITGA3
Description
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) [Source:HGNC Symbol;Acc:6139]
Coordinates
chr17:48165108-48167848:+
Coord C1 exon
chr17:48165108-48165233
Coord A exon
chr17:48165589-48165730
Coord C2 exon
chr17:48166474-48167848
Length
142 bp
Sequences
Splice sites
3' ss Seq
CCACCTCCTCCCCTCCGCAGTGC
3' ss Score
10.77
5' ss Seq
TGGGTAACA
5' ss Score
3.35
Exon sequences
Seq C1 exon
TTCTCTGTGGACATTGACTCGGAGCTGGTGGAGGAGCTGCCGGCCGAAATCGAGCTGTGGCTGGTGCTGGTGGCCGTGGGTGCAGGGCTGCTGCTGCTGGGGCTGATCATCCTCCTGCTGTGGAAG
Seq A exon
TGCGGCTTCTTCAAGCGAGCCCGCACTCGCGCCCTGTATGAAGCTAAGAGGCAGAAGGCGGAGATGAAGAGCCAGCCGTCAGAGACAGAGAGGCTGACCGACGACTACTGAGGGGGCAGCCCCCCGCCCCCGGCCCACCTGG
Seq C2 exon
TGTGACTTCTTTAAGCGGACCCGCTATTATCAGATCATGCCCAAGTACCACGCAGTGCGGATCCGGGAGGAGGAGCGCTACCCACCTCCAGGGAGCACCCTGCCCACCAAGAAGCACTGGGTGACCAGCTGGCAGACTCGGGACCAATACTACTGACGTCCTCCCTGATCCCACCCCCTCCTCCCCCAGTGTCCCCTTTCTTCCTATTTATCATAAGTTATGCCTCTGACAGTCCACAGGGGCCACCACCTTTGGCTGGTAGCAGCAGGCTCAGGCACATACACCTCGTCAAGAGCATGCACATGCTGTCTGGCCCTGGGGATCTTCCCACAGGAGGGCCAGCGCTGTGGACCTTACAACGCCGAGTGCACTGCATTCCTGTGCCCTAGATGCACGTGGGGCCCACTGCTCGTGGACTGTGCTGGTGCATCACGGATGGTGCATGGGCTCGCCGTGTCTCAGCCTCTGCCAGCGCCAAAACAAGCCAAAGAGCCTCCCAC
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000005884_CASSETTE5
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref, Alt. Stop)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.306 C2=0.000
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
NO
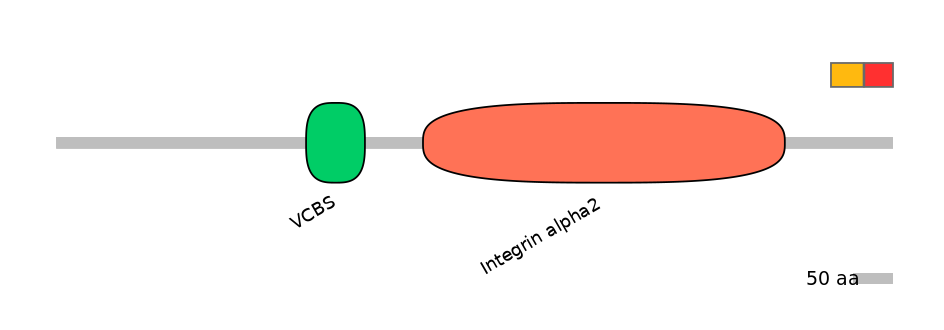
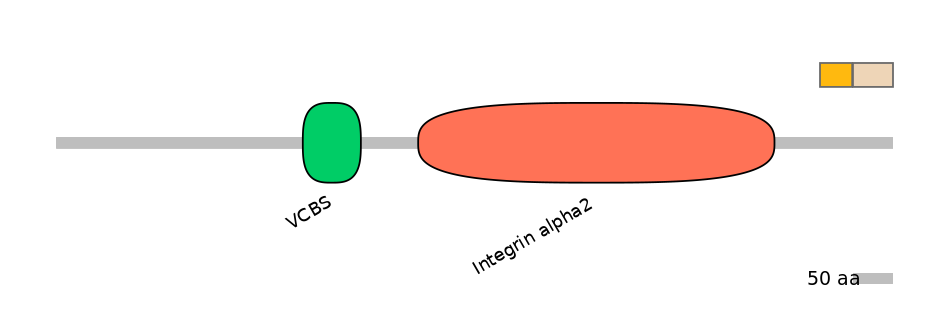
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GACATTGACTCGGAGCTGGTG
R:
GTCACCCAGTGCTTCTTGGTG
Band lengths:
242-384
Functional annotations
There are 2 annotated functions for this event
PMID: 10094488
This event
Two-yeast hybrid assay was used to examine interaction between various alpha integrins (including splice isoforms of integrin alpha 3 (alpha3A and alpha3B (these differ in presence of penultimae exon HsaEX0032352 (present in A, absent in B)), splice isoforms of integrin alpha 6 (alpha6A and alpha6B) (these differ in the presence of penultimate exon HsaEX0032370 (present in A, absent in B), and splice isoforms of integrin alpha 7 (alpha7A and alpha7B (these differ in presence of penultimate exon HsaEX0032377 (present in A, absent in B)) and four proteins, two of which are described as clones as they were identified in a cDNA screen (Mss4, BIN1, clone 63, and clone 80).
PMID: 11461117
Integrin alpha3beta1 can be alternatively spliced to generate alpha3A and alpha3B cytoplasmic domain variants that are conserved among vertebrates. To identify distinct functions of these variants, the authors transfected cells with intact alpha3 integrins or chimeric receptors. alpha3Abeta1 and alpha3Bbeta1 each localized to focal contacts in keratinocytes on an extracellular matrix rich in laminin-5, to which both are known to bind with high affinity. However, alpha3B accumulated intracellularly in keratinocytes on collagen, suggesting that laminin binding may stabilize alpha3Bbeta1 surface expression. Neither alpha3 cytoplasmic domain affected recruitment of chimeric alpha5 integrins to fibronectin-induced focal contacts, and either substituted for the alpha5 cytoplasmic domain in alpha5beta1-mediated cell migration. However, the alpha5/alpha3B chimera localized to cell-cell borders in MDCK or CHO cells to a lesser extent than did the alpha5/alpha3A chimera. To determine whether the alpha3 cytoplasmic domains conferred distinct localization to a nonintegrin protein, the authors transfected cells with interleukin-2 receptor (IL-2R) chimeras containing the alpha3 cytoplasmic domains. The IL-2R/alpha3A chimera was expressed efficiently on the cell surface, while the IL-2R/alpha3B chimera accumulated intracellularly. These findings suggest that the alpha3B cytoplasmic domain harbors a retention signal that is regulated in an intact integrin and can alter cell surface expression and distribution of alpha3beta1. Note, quite a lot of the results are of chimeric proteins.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)