HsaEX0032377 @ hg19
Exon Skipping
Gene
ENSG00000135424 | ITGA7
Description
integrin, alpha 7 [Source:HGNC Symbol;Acc:6143]
Coordinates
chr12:56078352-56081880:-
Coord C1 exon
chr12:56081755-56081880
Coord A exon
chr12:56079986-56080098
Coord C2 exon
chr12:56078352-56079072
Length
113 bp
Sequences
Splice sites
3' ss Seq
CTTCTCCCTCGACATCCTAGTGT
3' ss Score
5.21
5' ss Seq
GGGGTAACT
5' ss Score
4.69
Exon sequences
Seq C1 exon
ATCCCAGTGATGGTATACTTGGACCCCATGGCTGTGGTGGCAGAAGGAGTGCCCTGGTGGGTCATCCTCCTGGCTGTACTGGCTGGGCTGCTGGTGCTAGCACTGCTGGTGCTGCTCCTGTGGAAG
Seq A exon
TGTGGCTTCTTCCATCGGAGCAGCCAGAGCTCATCTTTTCCCACCAACTATCACCGGGCCTGTCTGGCTGTGCAGCCTTCAGCCATGGAAGTTGGGGGTCCAGGGACTGTGGG
Seq C2 exon
ATGGGATTCTTCAAACGGGCGAAGCACCCCGAGGCCACCGTGCCCCAGTACCATGCGGTGAAGATTCCTCGGGAAGACCGACAGCAGTTCAAGGAGGAGAAGACGGGCACCATCCTGAGGAACAACTGGGGCAGCCCCCGGCGGGAGGGCCCGGATGCACACCCCATCCTGGCTGCTGACGGGCATCCCGAGCTGGGCCCCGATGGGCATCCAGGGCCAGGCACCGCCTAGGTTCCCATGTCCCAGCCTGGCCTGTGGCTGCCCTCCATCCCTTCCCCAGAGATGGCTCCTTGGGATGAAGAGGGTAGAGTGGGCTGCTGGTGTCGCATCAAGATTTGGCAGGATCGGCTTCCTCAGGGGCACAGACCTCTCCCACCCACAAGAACTCCTCCCACCCAACTTCCCCTTAGAGTGCTGTGAGATGAGAGTGGGTAAATCAGGGACAGGGCCATGGGGTAGGGTGAGAAGGGCAGGGGTGTCCTGATGCAAAGGTGGGGAGA
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000135424-'32-33,'32-32,33-33
Average complexity
S
Mappability confidence:
100%=100=100%
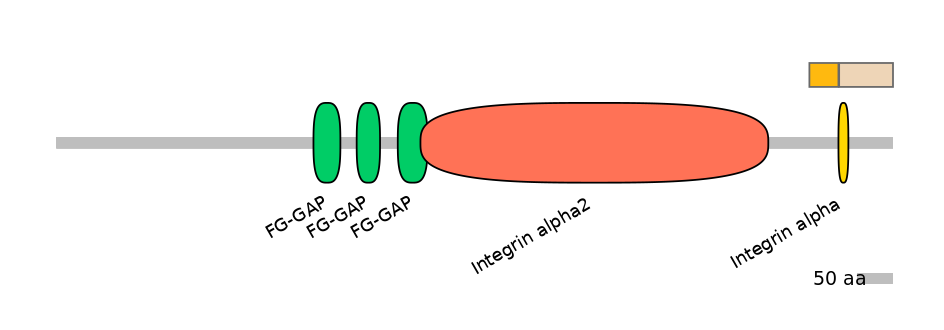
Protein Impact
Alternative protein isoforms (No Ref, Alt. Stop)
No structure available
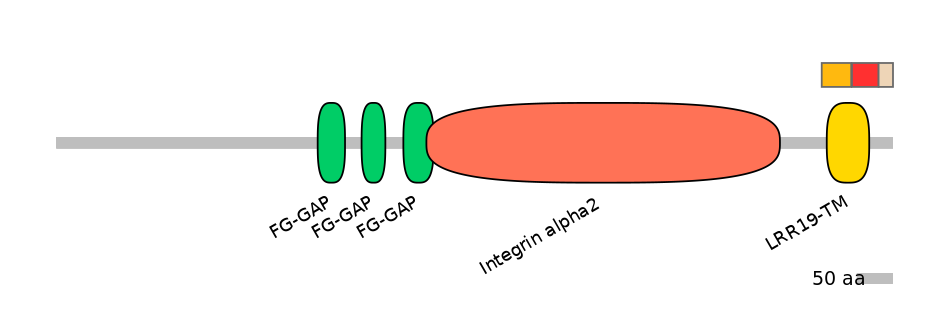
Features
Disorder rate (Iupred):
C1=0.000 A=0.053 C2=0.708
Domain overlap (PFAM):
C1:
PF0035715=Integrin_alpha=PU(0.1=0.0)
A:
PF151761=LRR19-TM=PD(40.0=63.2)
C2:
PF0035715=Integrin_alpha=PD(86.7=16.9)
Associated events
Other assemblies
Conservation
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ATACTTGGACCCCATGGCTGT
R:
CCCAGTTGTTCCTCAGGATGG
Band lengths:
242-355
Functional annotations
There are 2 annotated functions for this event
PMID: 10094488
This event
Two-yeast hybrid assay was used to examine interaction between various alpha integrins (including splice isoforms of integrin alpha 3 (alpha3A and alpha3B (these differ in presence of penultimae exon HsaEX0032352 (present in A, absent in B)), splice isoforms of integrin alpha 6 (alpha6A and alpha6B) (these differ in the presence of penultimate exon HsaEX0032370 (present in A, absent in B), and splice isoforms of integrin alpha 7 (alpha7A and alpha7B (these differ in presence of penultimate exon HsaEX0032377 (present in A, absent in B)) and four proteins, two of which are described as clones as they were identified in a cDNA screen (Mss4, BIN1, clone 63, and clone 80).
PMID: 10694445
This event
HEK293 overexpressing integrin alpha 7 isoforms alpha7A (contains penultimate exon (HsaEX0032377)) or alpha7B (lacks penultimate exon) are shown to adhere to similar extends to laminin-1, a fragment of laminin-1, and laminin-2. Migration across surfaces with above mentioned laminins is also similar. Overexpression of either integrin alpha 7 isoform negatively affects surface expression other beta1-integrin associated alpha integrin chains (alpha1, alpha3, alpha5, and alpha6). Overexpression also lead to reduced adhesiveness for both isoforms. Note, while they state that the isoforms behave similarly, I do think that their data points toward differences.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)