HsaEX0045708 @ hg19
Exon Skipping
Gene
ENSG00000007372 | PAX6
Description
paired box 6 [Source:HGNC Symbol;Acc:8620]
Coordinates
chr11:31823109-31824382:-
Coord C1 exon
chr11:31824252-31824382
Coord A exon
chr11:31823419-31823460
Coord C2 exon
chr11:31823109-31823324
Length
42 bp
Sequences
Splice sites
3' ss Seq
GGTATATTTTTGTGTTATAGACC
3' ss Score
6.86
5' ss Seq
AACGTAAGC
5' ss Score
7.35
Exon sequences
Seq C1 exon
GTCACAGCGGAGTGAATCAGCTCGGTGGTGTCTTTGTCAACGGGCGGCCACTGCCGGACTCCACCCGGCAGAAGATTGTAGAGCTAGCTCACAGCGGGGCCCGGCCGTGCGACATTTCCCGAATTCTGCAG
Seq A exon
ACCCATGCAGATGCAAAAGTCCAAGTGCTGGACAATCAAAAC
Seq C2 exon
GTGTCCAACGGATGTGTGAGTAAAATTCTGGGCAGGTATTACGAGACTGGCTCCATCAGACCCAGGGCAATCGGTGGTAGTAAACCGAGAGTAGCGACTCCAGAAGTTGTAAGCAAAATAGCCCAGTATAAGCGGGAGTGCCCGTCCATCTTTGCTTGGGAAATCCGAGACAGATTACTGTCCGAGGGGGTCTGTACCAACGATAACATACCAAGC
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000007372-'10-19,'10-18,11-19
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
Show PDB structure
Features
Disorder rate (disopred):
C1=0.101 A=0.000 C2=0.015
Domain overlap (PFAM):
C1:
PF0029213=PAX=PU(34.4=97.7)
A:
PF0029213=PAX=FE(20.6=100)
C2:
PF0029213=PAX=PU(88.0=98.6)
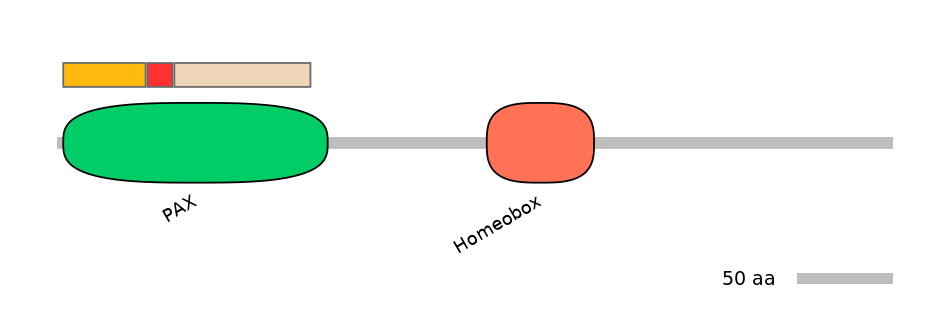
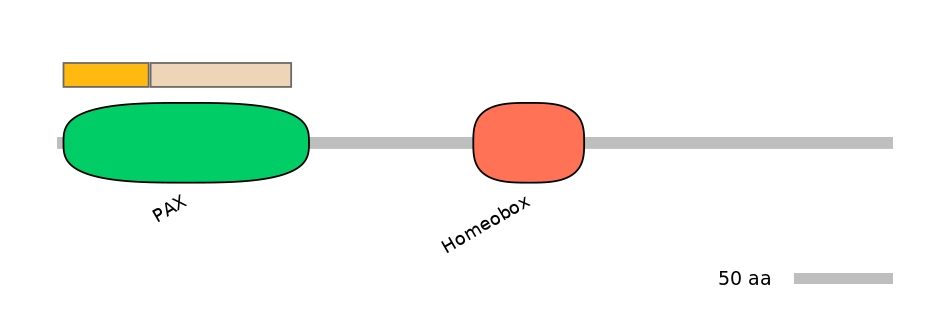
Other Inclusion Isoforms:
Other Skipping Isoforms:
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GAGTGAATCAGCTCGGTGGTG
R:
ACTCACACATCCGTTGGACAC
Band lengths:
143-185
Functional annotations
There are 8 annotated functions for this event
PMID: 7958875
This event
Inclusion isoform exhibits unique DNA-binding properties in its paired domain. Unlike other paired domains, which bind DNA predominantly by their amino termini, the extended Pax6 paired domain interacts with DNA exclusively through its carboxyl terminus. Thus, the insertion acts as a molecular toggle to unmask the DNA-binding potential of the carboxyl terminus.
PMID: 8640214
This event
The two PAX6 isoforms cooperatively act in the development of the posterior segment of the eye in humans
PMID: 15677484
This event
Overexpression of either human isoform in early stage chicken embryos increases retinal cell proliferation. -5a overexpression in later stage embryos leads to slight retinal overgrowth while +5a overexpression leads to extreme retinal overgrowth.
PMID: 26899008
This event
Structural variations in the PAI domain define two major PAX6 isoforms, isoform-a (PAX6-a) and isoform-b (PAX6-b). The key difference is that the PAI domain of PAX6-b possesses an extra exon 5a. To reveal the function of the two PAX6 isofoms in the corneal epithelium, PAX6 isoforms, along with reprogramming factors, were transduced into human non-ocular epithelial cells. Herein, the authors show that the two PAX6 isoforms differentially and cooperatively regulate the expression of genes specific to the structure and functions of the corneal epithelium, particularly keratin 3 (KRT3) and keratin 12 (KRT12). PAX6 isoform-a induced KRT3 expression by targeting its upstream region. KLF4 enhanced this induction. A combination of PAX6 isoform-b, KLF4, and OCT4 induced KRT12 expression.
PMID: 16725027
Both isoforms caused an up-regulation of endogenous Pax6 expression in cells with (Neuro2A) or without (NIH3T3) constitutive Pax6 expression. Western blots showed that cells stably transfected with constructs expressing either Pax6 or Pax6(5a) contained raised levels of both Pax6 and Pax6(5a). The overexpression of either isoform in neuroblastoma (Neuro2A) cell lines also promoted morphological change and an increase in beta-III-tubulin expression, indicating an increase in neurogenesis.
PMID: 22384097
A change in morphology of the stably transfected Pax6 and Pax6(5a) cells was observed, and the Pax6 expressing cells were shown to have increased proliferation and migration capacities. Further differences in functions during development are suggested by the fact that mutations in the 14 amino acid insertion for Pax6(5a) give a slightly different eye phenotype than the one described for Pax6.
PMID: 22384097
The Pax6(5a) protein differs from Pax6 by having a 14 amino acid insert in the paired domain, causing the two proteins to have different DNA binding specificities. Several genes are differentially regulated by each isoform, the majority of which were associated with the extracellular region. Verified by qPCR: Ncam1, Ngef, Sphk1, Dkk3 and Crtap for Pax6(5a); Tgfbi, Vegfa, EphB2, Klk8 and Edn1 for Pax6 (skipping). Nbl1, Ngfb and seven genes encoding different glycosyl transferases appeared to be regulated by both. Direct binding to the promoters of Crtap, Ctgf, Edn1, Dkk3, Pdgfb and Ngef was verified by ChIP.
PMID: 25029272
[Negative results]. By the use of bioinformatics and electrophoretic mobility shift assay (EMSA) the authors have mapped a functional Pax6 binding site in the mouse Dkk3 promoter. The minimal Dkk3 promoter fragment required for transcriptional activation by Pax6 and Pax6(5a) was a 200 bp region just upstream of the transcriptional start site.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)