HsaEX6018373 @ hg38
Exon Skipping
Gene
ENSG00000115414 | FN1
Description
fibronectin 1 [Source:HGNC Symbol;Acc:HGNC:3778]
Coordinates
chr2:215383328-215384976:-
Coord C1 exon
chr2:215384860-215384976
Coord A exon
chr2:215384020-215384184
Coord C2 exon
chr2:215383328-215383483
Length
165 bp
Sequences
Splice sites
3' ss Seq
ACTAACCCTTTTTCATACAGGAG
3' ss Score
8.16
5' ss Seq
CAGGTACAA
5' ss Score
7.09
Exon sequences
Seq C1 exon
TTTCTGATGTTCCGAGGGACCTGGAAGTTGTTGCTGCGACCCCCACCAGCCTACTGATCAGCTGGGATGCTCCTGCTGTCACAGTGAGATATTACAGGATCACTTACGGAGAGACAG
Seq A exon
GAGGAAATAGCCCTGTCCAGGAGTTCACTGTGCCTGGGAGCAAGTCTACAGCTACCATCAGCGGCCTTAAACCTGGAGTTGATTATACCATCACTGTGTATGCTGTCACTGGCCGTGGAGACAGCCCCGCAAGCAGCAAGCCAATTTCCATTAATTACCGAACAG
Seq C2 exon
AAATTGACAAACCATCCCAGATGCAAGTGACCGATGTTCAGGACAACAGCATTAGTGTCAAGTGGCTGCCTTCAAGTTCCCCTGTTACTGGTTACAGAGTAACCACCACTCCCAAAAATGGACCAGGACCAACAAAAACTAAAACTGCAGGTCCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000115414-'76-93,'76-89,79-93
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.075 A=0.598 C2=0.685
Domain overlap (PFAM):
C1:
PF0004116=fn3=PU(45.1=92.5)
A:
PF0004116=fn3=PD(53.7=78.6)
C2:
PF0004116=fn3=PU(61.7=94.3)
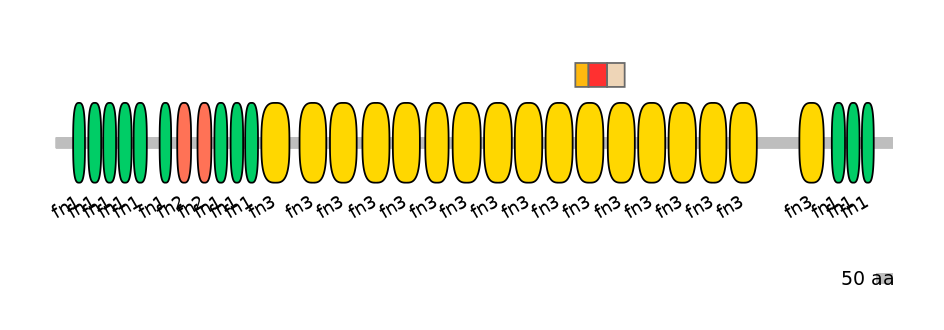
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TCTGATGTTCCGAGGGACCTG
R:
TGGTCCTGGTCCATTTTTGGG
Band lengths:
246-411
Functional annotations
There are 4 annotated functions for this event
PMID: 6325925
Encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: cross linking study, overlapping binding peptide, x-ray crystallography. ELM ID: ELMI000104; ELM sequence: RGD; Overlap: FULL
PMID: 19738201
Encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: cross linking study, overlapping binding peptide, x-ray crystallography. ELM ID: ELMI000104; ELM sequence: RGD; Overlap: FULL
PMID: 22451694
Encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: cross linking study, overlapping binding peptide, x-ray crystallography. ELM ID: ELMI000104; ELM sequence: RGD; Overlap: FULL
PMID: 9533887
Encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: structure based prediction. ELM ID: ELMI000105; ELM sequence: RGD; Overlap: FULL
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Autistic and control brains