HsaINT0041176 @ hg19
Intron Retention
Gene
ENSG00000117322 | CR2
Description
complement component (3d/Epstein Barr virus) receptor 2 [Source:HGNC Symbol;Acc:2336]
Coordinates
chr1:207643040-207644261:+
Coord C1 exon
chr1:207643040-207643447
Coord A exon
chr1:207643448-207644084
Coord C2 exon
chr1:207644085-207644261
Length
637 bp
Sequences
Splice sites
5' ss Seq
AGGGTGAGT
5' ss Score
9.25
3' ss Seq
GTTTTTGTCCTTTCATTTAGAAT
3' ss Score
9.64
Exon sequences
Seq C1 exon
AAATTTTTTGCCCATCACCTCCCCCTATTCTCAATGGAAGACATATAGGCAACTCACTAGCAAATGTCTCATATGGAAGCATAGTCACTTACACTTGTGACCCGGACCCAGAGGAAGGAGTGAACTTCATCCTTATTGGAGAGAGCACTCTCCGTTGTACAGTTGATAGTCAGAAGACTGGGACCTGGAGTGGCCCTGCCCCACGCTGTGAACTTTCTACTTCTGCGGTTCAGTGTCCACATCCCCAGATCCTAAGAGGCCGAATGGTATCTGGGCAGAAAGATCGATATACCTATAACGACACTGTGATATTTGCTTGCATGTTTGGCTTCACCTTGAAGGGCAGCAAGCAAATCCGATGCAATGCCCAAGGCACATGGGAGCCATCTGCACCAGTCTGTGAAAAGG
Seq A exon
GTGAGTGTTCCGGTACTCAGAAAAGGTGCTTCTGATTCGTTTCTGAAAAATTAGAAGAAGGGGTTGTGGGCTTTAGGTAGGGCCTTGTCCAGTTTATACTTCCCTCAAATCTACTACATCTAATCAATATAAATTTTGTAGAGGGCATTCTTATCACTAGCCCCCCACCATTGTTTTATTTTGTGGGAATATGCTTGGAAAAAATGTTAGGAATCACTAAGTTTCTCATTTCTATAGGGGAAAAAATGAGGAGAAAAATGCTTGTTTGTCTTAATAGTGACTTCTTAAAAGAGAAGTCATTCAAGCCCTCATTCCTAGGGATATATCAGAATCTCCCATAAAAAACATACAAGATGATTCCTTATGAAGGGAAGAGGCAGGAGAGGAGTTGCAGAACCCAGTGGAAAGTGAACAACATCTGCAGCAGCCTCTGTGCAGAAAACAACAACAACAACAACAACAAAAGATTGGGAAACTGTGATCTAAAATTACCCAAAGCTGGTCTGCAACATATGTTCTGTATCATACAGCTGACGCCAGAGTGGAATTATAGCATGAATATCAATTTCTTTGGCTCAGTTTCTTTCTGTGGTTGTTTACTTAAGCAGTTATGTTTTGTTTTTGTCCTTTCATTTAG
Seq C2 exon
AATGCCAGGCCCCTCCTAACATCCTCAATGGGCAAAAGGAAGATAGACACATGGTCCGCTTTGACCCTGGAACATCTATAAAATATAGCTGTAACCCTGGCTATGTGCTGGTGGGAGAAGAATCCATACAGTGTACCTCTGAGGGGGTGTGGACACCCCCTGTACCCCAATGCAAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000117322-CR2:NM_001877:6
Average complexity
IR-S
Mappability confidence:
NA
Protein Impact
ORF disruption upon sequence inclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=NA C2=0.073
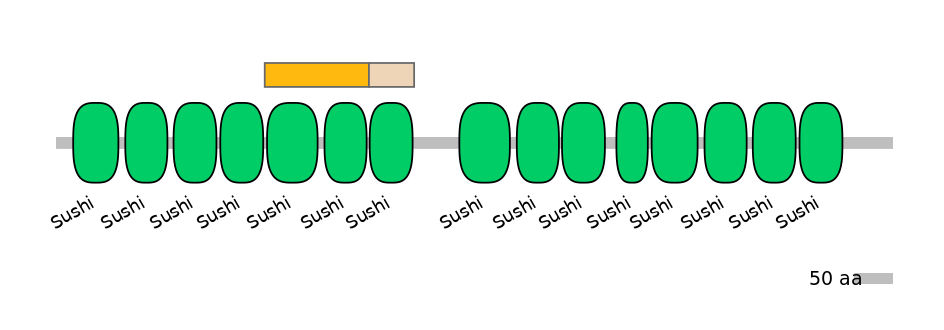
Domain overlap (PFAM):
C1:
PF0008415=Sushi=WD(100=48.9),PF0008415=Sushi=WD(100=40.9)
A:
NA
C2:
PF0008415=Sushi=WD(100=95.0)
Main Inclusion Isoform:
NA
Other Inclusion Isoforms:
NA
Associated events
Other assemblies
Conservation
Rat
(rn6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CCCAGATCCTAAGAGGCCGAA
R:
CTTTGCATTGGGGTACAGGGG
Band lengths:
342-979
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)