MmuALTA0008176-4/4 @ mm10
Alternative 3'ss
Gene
ENSMUSG00000026959 | Grin1
Description
glutamate receptor, ionotropic, NMDA1 (zeta 1) [Source:MGI Symbol;Acc:MGI:95819]
Coordinates
chr2:25292058-25295937:-
Coord C1 exon
chr2:25295792-25295937
Coord A exon
chr2:25292126-25292484
Coord C2 exon
chr2:25292058-25292125
Length
359 bp
Sequences
Splice sites
5' ss Seq
CAGGTAGGA
5' ss Score
9.79
3' ss Seq
TTGTTGGTTCTTATTTATAGAGC
3' ss Score
6.94
Exon sequences
Seq C1 exon
GGGTCTTCATGCTGGTGGCTGGAGGCATCGTAGCTGGGATCTTCCTCATTTTCATCGAGATCGCCTACAAGCGACACAAGGATGCCCGTAGGAAGCAGATGCAGCTGGCTTTTGCAGCCGTGAACGTGTGGAGGAAGAACCTGCAG
Seq A exon
AGCACCGGGGGTGGACGCGGCGCTTTGCAAAACCAAAAAGACACAGTGCTGCCGCGACGCGCTATTGAGAGGGAGGAGGGCCAGCTGCAGCTGTGTTCCCGTCATAGGGAGAGCTGAGACGCCCCGCCCGCCCTCCTCTGCCCCTCCCCCGCAGACAGACGCACGGGACAGCGGCCTGGCCCACGCAGAGCCCCGGAGCACGACGGGGTCGGGGGAGGAGCACCCCCAGCCTCCCCCAGGCCGTGCCCGCCTGCCCGCCGGTCGGCCGGCTGGCCGGTCCACCCTGTCCCGGCCCCGCGCGTGCCCCCGACGTCGGAGCTAACGGGCCGCCTTGTCTGTGTATTTCTATTTTACAGCAG
Seq C2 exon
TACCATCCCACTGATATCACGGGCCCGCTCAACCTCTCAGATCCCTCGGTCAGCACCGTGGTGTGAGG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000026959-32-40,32-39,32-38,32-37-4/4
Average complexity
Alt3
Mappability confidence:
NA
Protein Impact
Protein isoform when splice site is used (No Ref, Alt. Stop)
No structure available
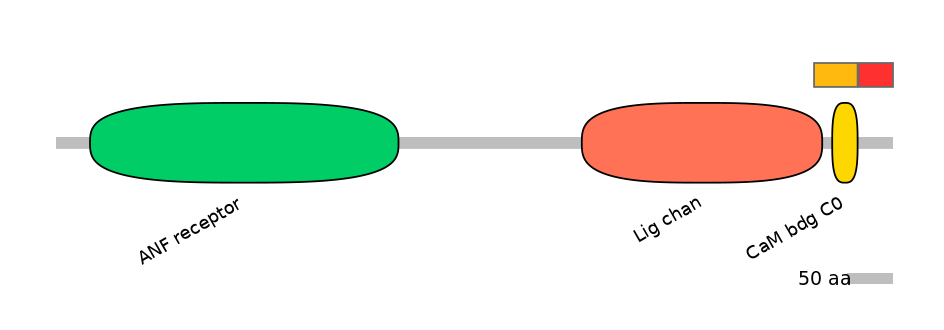
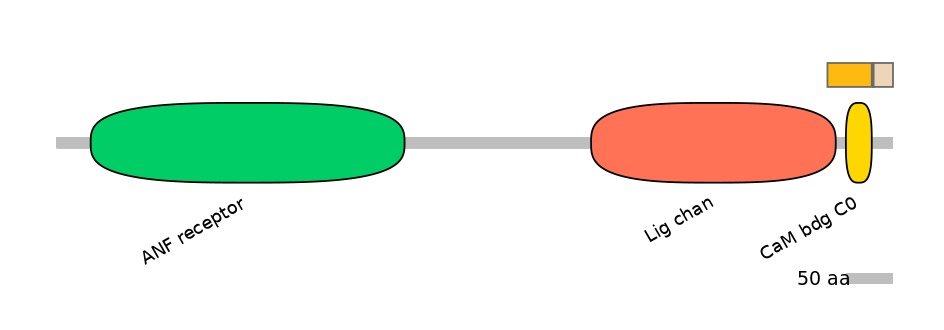
Features
Disorder rate (Iupred):
C1=0.005 A=NA C2=0.433
Domain overlap (PFAM):
C1:
PF0006021=Lig_chan=PD(3.4=18.4),PF105624=CaM_bdg_C0=WD(100=59.2)
A:
NA
C2:
NO
Associated events
Conservation
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GGTCTTCATGCTGGTGGCTG
R:
CTGACCGAGGGATCTGAGAGG
Band lengths:
198-557
Functional annotations
There are 2 annotated functions for this event
PMID: 10479681
This event
When NR1 splice variants were expressed in fibroblasts, the amount of NR1 molecules expressed on the cell surface varied among forms with different C-terminal cytoplasmic domains. The splice forms with the longest C-terminal cytoplasmic tail (NR1-1a and NR1-1b) showed the lowest amount of cell surface expression, and the splice forms with the shortest C-terminal cytoplasmic tail (NR1-4a and NR1-4b) showed the highest cell surface expression. Cells transfected with 1b, 2b, 3b, or 4b demonstrate different levels of [Ca2+] increase following glutamate stimulation. N-cassette: MmuEX0021891??, C1: MmuEX0021891, C2-C2': MmuALTA0008176.
PMID: 34187890
This event
The GluN1 subunit contains eight alternatively spliced isoforms produced by including or excluding the N1 and the C1, C2, or C2' polypeptide cassettes. Whether GluN1 alternative splicing affects nonionotropic signaling by NMDARs is a major outstanding question. Here, the authors discovered that glycine priming of recombinant NMDARs critically depends on GluN1 isoforms lacking the N1 cassette; glycine priming is blocked in splice variants containing N1. On the other hand, the C-terminal cassettes-C1, C2, or C2'-each permit glycine signaling. In wild-type mice, the authors found glycine-induced nonionotropic signaling at synaptic NMDARs in CA1 hippocampal pyramidal neurons. This nonionotropic signaling by glycine to synaptic NMDARs was prevented in mice the authors engineered, such that GluN1 obligatorily contained N1. The authors discovered in wild-type mice that, in contrast to pyramidal neurons, synaptic NMDARs in CA1 inhibitory interneurons were resistant to glycine priming. But the authors recapitulated glycine priming in inhibitory interneurons in mice engineered such that GluN1 obligatorily lacked the N1 cassette. Ex 5: MmuEX0021893, C-term cassettes: MmuALTA0008176 and MmuEX0021891.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Ribosome-engaged transcriptomes of neuronal types
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types