MmuALTA1002178-2/2 @ mm10
Alternative 3'ss
Gene
ENSMUSG00000004872 | Pax3
Description
paired box 3 [Source:MGI Symbol;Acc:MGI:97487]
Coordinates
chr1:78193740-78195486:-
Coord C1 exon
chr1:78195251-78195486
Coord A exon
chr1:78193867-78193869
Coord C2 exon
chr1:78193740-78193866
Length
3 bp
Sequences
Splice sites
5' ss Seq
AAGGTGAGC
5' ss Score
9.6
3' ss Seq
ATCCGCCTATTCTCTTAAAGCAG
3' ss Score
6.55
Exon sequences
Seq C1 exon
TGTCCACCCCTCTTGGCCAGGGCCGAGTCAACCAGCTCGGAGGAGTATTTATCAACGGCAGGCCTCTGCCCAACCATATCCGCCACAAGATAGTGGAGATGGCCCACCATGGCATTCGGCCTTGCGTCATTTCTCGCCAGCTTCGCGTGTCCCATGGTTGCGTCTCTAAGATCCTGTGCAGGTACCAGGAGACAGGCTCCATCCGACCTGGTGCCATCGGCGGCAGCAAACCCAAG
Seq A exon
CAG
Seq C2 exon
GTGACAACGCCTGACGTGGAGAAGAAAATTGAGGAATACAAAAGAGAGAACCCGGGCATGTTTAGCTGGGAAATCAGAGACAAATTGCTCAAGGACGCTGTCTGTGATCGGAACACTGTGCCCTCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000004872-1-2,1-1-2/2
Average complexity
Alt3
Mappability confidence:
NA
Protein Impact
Protein isoform when splice site is used (Ref)
No structure available
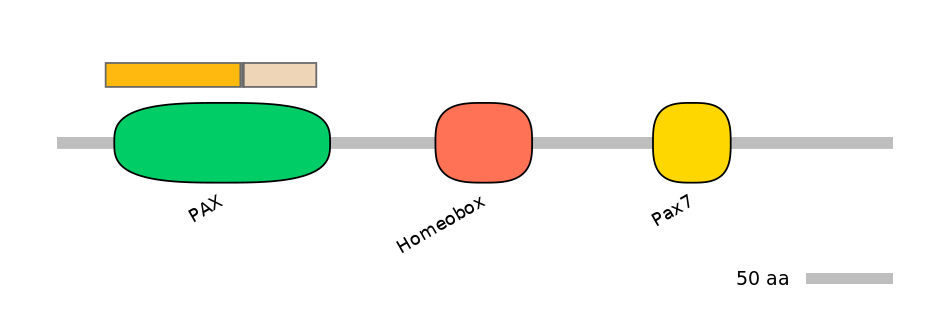
Features
Disorder rate (disopred):
C1=0.215 A=NA C2=0.000
Domain overlap (PFAM):
C1:
PF0029213=PAX=PU(57.9=92.4)
A:
NA
C2:
PF0029213=PAX=FE(34.1=100)
Main Skipping Isoform:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GAGACAGGCTCCATCCGACC
R:
ATGCCCGGGTTCTCTCTTTTG
Band lengths:
107-110
Functional annotations
There are 2 annotated functions for this event
PMID: 15688409
DNA binding analysis demonstrated that PAX3 Q- (taking internal NAGNAG) isoforms exhibit higher affinity than corresponding Q+ isoforms for class I sites and no difference for class II sites. This results in different transcriptional activity.
PMID: 15688409
DNA binding analysis demonstrated that PAX3 Q- (taking internal NAGNAG) isoforms exhibit higher affinity than corresponding Q+ isoforms for class I sites and no difference for class II sites. This results in different transcriptional activity.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Ribosome-engaged transcriptomes of neuronal types
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types