MmuEX0024626 @ mm9
Exon Skipping
Gene
ENSMUSG00000025348 | Itga7
Description
integrin alpha 7 [Source:MGI Symbol;Acc:MGI:102700]
Coordinates
chr10:128377738-128380071:+
Coord C1 exon
chr10:128377738-128377993
Coord A exon
chr10:128378878-128378997
Coord C2 exon
chr10:128379864-128380071
Length
120 bp
Sequences
Splice sites
3' ss Seq
CCCCGACCTGCTCCCCACAGGGT
3' ss Score
9.7
5' ss Seq
TAGGTTTGT
5' ss Score
4.07
Exon sequences
Seq C1 exon
ACGTGTGCACACCGATATGAGTCTCGACAGAGAGTGGACCAGGCTTTGGAGACTCGGGATGTGATTGGCCGCTGCTTTGTGCTAAGCCAGGACCTGGCCATCCGTGATGAGCTGGATGGTGGGGAGTGGAAGTTCTGTGAGGGGCGCCCCCAGGGCCATGAACAGTTTGGGTTCTGTCAGCAGGGCACAGCTGCCACCTTCTCCCCTGACAGTCACTACCTCGTCTTTGGGGCTCCAGGAACCTATAACTGGAAGG
Seq A exon
GGTTGCTTTTTGTGACCAACATTGATAGCTCAGACCCTGACCAGCTGGTGTATAAAACTTTGGACCCTGCTGACCGGCTCACAGGACCAGCCGGAGACTTGACCTTGAATAGCTATTTAG
Seq C2 exon
GTTTCTCCATCGATTCTGGGAAGGGTCTTATGCGCTCAGAAGAGCTGAGTTTTGTGGCAGGGGCCCCACGTGCCAACCACAAAGGGGCTGTGGTCATTCTGCGCAAGGATAGCGCCAGCCGCTTGATACCTGAGGTTGTGCTGTCTGGGGAGCGCCTGACCTCTGGCTTTGGCTACTCACTGGCTGTGACTGATCTCAACAATGATGG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000025348_MULTIEX1-2/2=C1-C2
Average complexity
ME(1-2[100=100])
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.012 A=0.061 C2=0.000
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
PF135171=VCBS=PU(6.8=7.1)
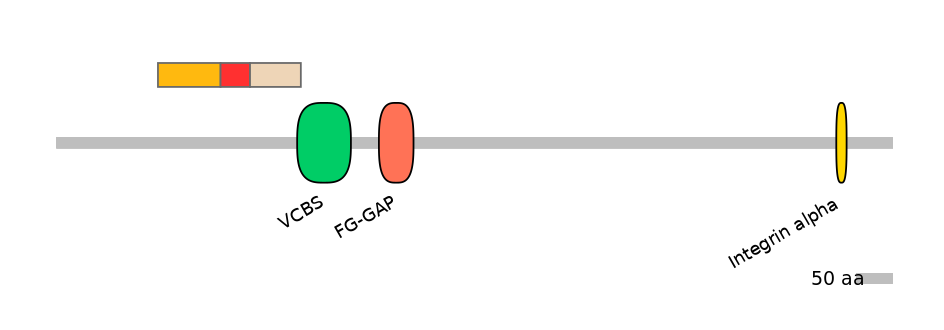
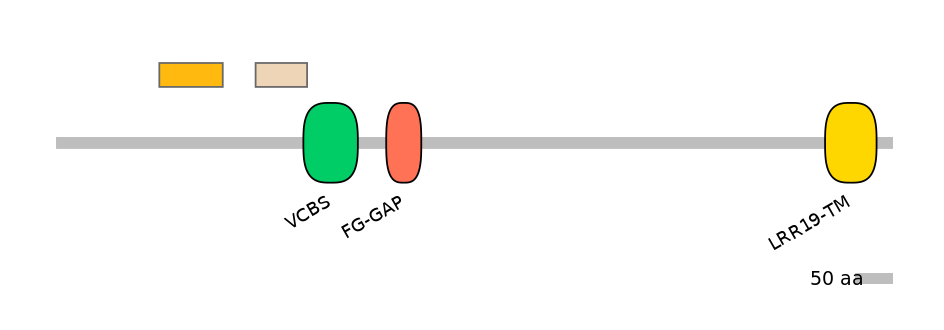
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Chicken
(galGal4)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CACCTTCTCCCCTGACAGTCA
R:
CCAGTGAGTAGCCAAAGCCAG
Band lengths:
245-365
Functional annotations
There are 3 annotated functions for this event
PMID: 9307969
The expression pattern of the laminin-binding alpha 7 beta 1 integrin is developmentally regulated in skeletal, cardiac, and smooth muscle. The X1/X2 alternative splicing (exon X1 is encoded by HsaEX0032381, exon X2 is encoded by HsaEX0032382. Both are cassette exons) in the extracellular domain of alpha 7 is found in the variable region between conserved alpha-chain homology repeat domains III and IV, a site implicated in ligand binding. To assess differences in X1/X2 isoform activity, the authors generated MCF-7 cell lines transfected with alpha 7-X1/X2 cDNAs. Transfectants expressing the alpha 7-X2 variant adhered rapidly to laminin 1, whereas those expressing alpha 7-X1 failed to attach. That alpha 7-X1 exists in an inactive state was established in assays using an activating beta 1 antibody that induced X1-dependent cell adhesion and spreading. Furthermore, the activation of alpha 7-X1 was cell type specific, and when expressed in HT1080 cells, the integrin was converted into a fully functional receptor capable of promoting adhesion.
PMID: 10694445
Overexpression of either of two extracellular integrin alpha 7 splice variants a7X1 (contains cassette exon X1 (HsaEX0032381), lacks cassette exon X2 (HsaEX0032382)) and a7X2 (lacks X1, contains X2) in HEK293 cells conferred different motilities on laminin isoforms: Whereas a7X2B promoted cell migration on both laminin-1 and laminin-2, a7X1B supported motility only on laminin-2 and not on laminin-1, although both X1 and X2 splice variants revealed similar adhesion rates to laminin-1 and -2.
PMID: 11744715
The integrin alpha7beta1 occurs in several cytoplasmic (alpha7A, alpha7B) and extracellular splice variants (alpha7X1 (contains exon X1 (HsaEX0032381), lacks exon X2 (HsaEX0032382)), and alpha7X2 (lacks X1, contains X2)), which are differentially expressed during development of skeletal and heart muscle. The extracellular variants result from the alternative splicing of exons X1 and X2, corresponding to a segment within the putative ligand binding domain. To study the specificity and affinity of the X1/X2 variants to different laminin isoforms, soluble alpha7beta1 complexes were prepared by recombinant coexpression of the extracellular domains of the alpha- and beta-subunits. The binding of these complexes to purified ligands was measured by solid phase binding assays. Surprisingly, the alternative splice variants revealed different and specific affinities to different laminin isoforms. While the alpha7X2 variant bound much more strongly to laminin-1 than the alpha7X1 variant, the latter showed a high affinity binding to laminins-8 and -10/11. Laminin-2, the major laminin isoform in skeletal muscle, was recognized by both variants, whereas none of the two variants were able to interact with laminin-5. A specific blocking antibody inhibited the binding of both variants to all laminins tested, indicating the involvement of common epitopes in alpha7X1beta1 and alpha7X2beta1.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types
Other AS DBs: