MmuEX6034816 @ mm9
Exon Skipping
Gene
ENSMUSG00000031481 | Tpte
Description
transmembrane phosphatase with tensin homology [Source:MGI Symbol;Acc:MGI:2446460]
Coordinates
chr8:23416310-23421799:+
Coord C1 exon
chr8:23416310-23416366
Coord A exon
chr8:23417408-23417458
Coord C2 exon
chr8:23421752-23421799
Length
51 bp
Sequences
Splice sites
3' ss Seq
TGTACTTTTCAATAATTTAGAGC
3' ss Score
6.32
5' ss Seq
CAGGTGAGT
5' ss Score
10.67
Exon sequences
Seq C1 exon
TTTGTCATATCCAGATGAAATAAAATCAGCAAGTTATGCGGACCCCATCAGTACAAA
Seq A exon
AGCATATACAAATGATTCGTCAGTATACGACCCTGGGGGAGCTTCGAGCAG
Seq C2 exon
CACAACCCTGTATGAACTGAATTCCTTGAGTGAAGTATCGAAAGAAAT
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000031481-'7-7,'7-6,8-7=AN
Average complexity
A_C2
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
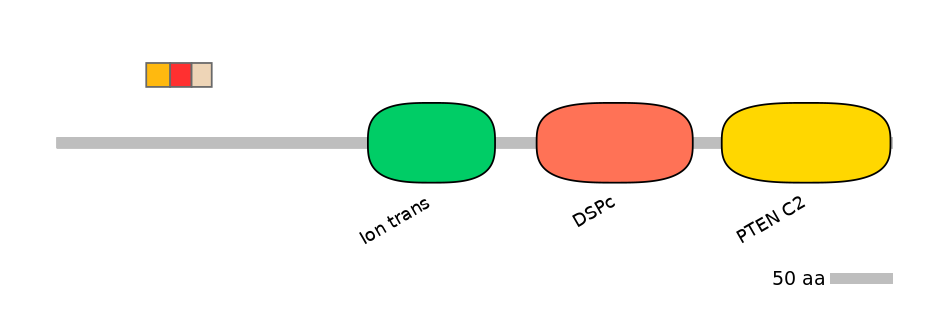
Features
Disorder rate (Iupred):
C1=0.050 A=0.417 C2=0.000
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
NO
Main Skipping Isoform:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TCCAGATGAAATAAAATCAGCAAGT
R:
TCTTTCGATACTTCACTCAAGGA
Band lengths:
93-144
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
Other AS DBs: