MmuINT0110513 @ mm9
Intron Retention
Gene
ENSMUSG00000032292 | Nr2e3
Description
nuclear receptor subfamily 2, group E, member 3 [Source:MGI Symbol;Acc:MGI:1346317]
Coordinates
chr9:59790578-59794874:-
Coord C1 exon
chr9:59794769-59794874
Coord A exon
chr9:59791327-59794768
Coord C2 exon
chr9:59790578-59791326
Length
3442 bp
Sequences
Splice sites
5' ss Seq
GAGGTGACA
5' ss Score
3.85
3' ss Seq
GTGTCCTTTCTCCTGTTCAGGTT
3' ss Score
11.34
Exon sequences
Seq C1 exon
AAACACGAGGCCTGAAGGATCCTGAGCACGTGGAGGCTTTGCAGGACCAGTCCCAGGTGATGCTAAGCCAGCATAGCAAGGCTCACCACCCCAGCCAGCCTGTGAG
Seq A exon
GTGACAGCAATGACCCACCTGCCCACCTGCCCTCCCCACTTGCCTCTCCCATACCATCTCTGGGCTTCCCTGCTTAGACAGGATAAAGGGCTCAGTCAGCCACAGCTATCCAACATCTCCTGTGGACCAGGCTCAGTGCCAGGCCCAGGAGGACATCAAGAGACAAAGCCTACGGGGATGTTCATTATGGTAGCCACATCAAAGACAAGAACCATCCTCATCAGGTGTAGAGTGGCTTGTGTCTGAAAGGGTAGCAAATGGGCAGGGAGGTGGTGGCGCATGCCTTTCATCCTAGCACTCGGGAGGTAGAAGCAGGCAGATAACTGAGTCTGAGGCCAGCCAGAGCAAGTTCAAGGACAGCCTGGCTACACAAAGAAACCCTCTCTCCAAAAACAAACAAAAAATTACTTTTAAAAGATAGGATGTCAGTATAGTCTTAACTGGTCTCAAAGTCAGTACTGGGCTTAAAGGTGTTTCCTACCAGCCTTTCACAGGGACCATGTGTGTCCATGACAGGATAGGATCTAAACAGAAGAGAAGAGTATTATCTATGAAGGTAGAGAGAGCCCAGCAAAAATGTGTTCTTAGACAGGAGTCAGGTATGGGAGGTTCAGAAGATTCTTGGTGGTAGGAAGGTGGGAGATGAGCCAGAGAGTTACCTTGGAACCAGTCGGACTCGGACTAGAGCTCAGAATGCCTAGTCAGATGAGAGAGGTGAGGAGAACCCCTGAGGTTCTGTGAGAGGTGGCCTTCATGCGCGGGACTGAACCCCATGCAGACTGGTGGGCAGTATGCACCAAGGGCAGAGCAGTGACTTCCCTGTCCGGGAGATTCTGTGCCTTGGGGAGTGGCTATAGGAAGCAGGGAAACAGACTGCAGTATACAACTCTGAGAACTGGAAAGGGACAGGTTTGGCATTCGGCTAAAGGGAAAACACACACACACACACACACACACAGGCAAAGAGATGAATTTGGAAGCAGAGAGAGTTCAAAACTTAAAAGGAAAGTTGAGTTTAGAAGGAACTTGCCAAGCTTAAGTGTGGAAGTCTCGCTGTGGAGATGTATGGAAAAGCCAGGCATGAAGGAACATGGCTATGGCCAGCTAGTGCAGGCTGCATGGAGGGGGTAGACACAGGTGGAGGATCTCAACGTAGTCTAGCACCAGGTCTTACACTGAAGCACCAGGTAGTTTATACCACCAAGCCAGGCCTTTGGACTATTCTAGACCACACTGTCTGCCACCTTCCTGAAGAACTGTGGTGACATAATCATCTTTAGTCACCTTGCAGGTAGGCAAGCTCTGCACCACATGGTACAGGGCCAGAGGGAGAACAGTGAAGGTTTGGCATAGTTTCACATGTGTGGTCATCACTGACATTCAGAAACCAAGGCTCTTCCTGGGTCAAGTTTGCTCTGGGACCAGCCCTCAGGGGCACCCTGGCCTGTTGACTGTGTATTATTGCAAAGGTACTGGGGAAGACTTCCTATGGAGGGGCAGGAGAAACTTGAGCCACCCAGAGTCTGCCAAACAAGCATGAAGACCTGCCAGGCATGGTGGCAGGGATCTGTCACGCCAGCACTGATCTGGGTGGAGACAGGTGGATGCAGCTTAGCTGAACTGACGAGTTCCAGGTTCAGTTGAGACAACTCTTCTCAATAAACAGGGAGGAAGGGCCAGGACTAAGAGTATTTGCTGTGCACGCATGAGGATAGGGGTTCAAATCCCCAGCACCCATGAGAGAGAGCTGAAAATGATGGGGATAGCAGGTTAAATAAATAAAAACCAGTGCTGGGCAGCAGAGCCAGGCAGATCCTGGGAGCACTCTGTCCTGCTAGCCTAGCCGAAATAACAAGGCTCAGGTTCAGAGAGAAACCTTGTCTCGAGGGAAGAAAGCAGAGATGTATGGGATGTCTGGTGTCTTCCTCTGACCTCATATATACACCAGCACACATGTGTGTCCATATATGTGGGTGCATACACACACACACACTAAAAATAAAGTGGACAGTGATTGAAGACTAGAATGCGACCTCTGGCTCTCCACACAGACTCTCCAGGTCACTTCTGCAGCTTAATACGTATGTCAGGTGGGACTGGGTGTCATGGCAATGTACTTTAGCCCAGCCTACAGAGAGAGAGAGAGAGATGCTAAAGCCCAGTTCCACTAATTATACCCAGCCTTGTATGGTCTGTATGGTTTGCTCCAATACATTGTTTTGAGACTAAGTTCAATGGACTCCAAGCTCAGTCCTGAATTCACTACGCCAGTGGGGATAAGCTGAGCTTCCACCCTTCACTTCCAGAGCAAAAGAGCATATGCAGCCACATGGGGTCCATGTGCTGCTGGGGACAGAGCCTAGGGTCTGGTGCCGGCTGAGCAAGAAGGGAAGGTGGAGCAGGGCAGGGGTGGGGCGGGGGTGACTGCTTACATGCTGCTCATAAGAGAAAGGGAGGTTTTAGCTGGGCAAAGCCACTCACTAGGTCAGAGAGTAAATGGCTGGTTTCTCATAGTTCTCTAGGTGCAGTCAAGCCAGCAGCTGCCTCGTGGAGCTCAGCCTTCTTGGTGTGTGGGTGGGTGTGGGTGTGTGGGTGGGTTAGTGGGGGAAGCCTTAGGCCTTCTTCCCAGGGTCATCTGGTTCTGATTTTTTTTTTAATTTTTTGTACTGGGGACACCTGCTTTAGCAAATGGTTCTTACTTGAAACTAGAACGGGTTCTTTTTCTCTGGATATATCTCACTTAACTAACATCTCCCTTGACTTCGAATATTTTCAGTAGTAGTTCCTGTCATCAAAGGTGTTTCATGGACAGGAACTCATCTATGTCCATGTAGAACTAACGTGACTTAGTTATAGCACTCATGATCAATGAGCCTCCGGACAGGATTGCTACCAACGCCTCGCAGGCTGGGAGAGAGCTGTACAAGTATGGAGACTGGAGCTCAGATCCCTGGGACGGGCACAAAAGCTGGCAGGCATAGCAGGCCTGCCATTGCAGCACCTGGGAGACTGACACAGGATCCCAGGGGCAAGTTAGCCAGAATTGGTGAGCTTCCAGGTCAGCTAGGGATCCAGCCTCAGTAAATAGAGAATGATCTAGAAAGACACTTGACCTCAACTTCCACCCAAACACATGAGCACTTGTACACACTTAGGGCACACAGACAGACATACAGACACACACACACACACACACACACACACACACACATCACATGCATGTGCAAAGAAACCTCTCATTTCTTAAATCTTGATGCATAGTGAACACTATTGAGTCCCTGTTTTCATCACGCTAATAACCAATGGGAAGGTGGACGTGAGTCTTCTAATTCAGGGGCTTTCCCTGTATTGAAATGCCCCTGGCTGGCCCTGGACAACTCCCTCCCCTCCCCCCACTGCCTTGCTCGGAGGTTGGCCTGACAGTGTCCTTTCTCCTGTTCAG
Seq C2 exon
GTTTGGGAAATTGCTCCTCCTGCTCCCATCTTTGAGGTTCCTCACGGCTGAGCGCATTGAGCTTCTCTTCTTCAGAAAGACCATAGGGAACACTCCGATGGAGAAGCTCCTGTGTGACATGTTCAAAAACTAGTTGGGAGTGCCAAGTGTCCACAGGCACCCAGGGGGGCAGCACATCTTAGAAGCTAAATAGTTCCCTGCCTTTCTCAGCCAGTAATTCCACATTCAGGTATTCCTACCTAGCAGAAATTTCTCCCAAAATATATTATTGGCATATTCATTGCCATCCTAATCTTAATACCCCTAACTCTGCTTGGGCAGTAGAATGCATGGATGCGTTGTTATATTCATAGGAGAAACAGCTTTGGCAAAAAATGAAATAAAAAATAAAAATGAACAGTAGGTCCATGAGACATGGAGACATTTCATAGGCTGGAAGTTGAACAAAAGCCAGGCACGTTTATCCTGTGTTCAAGACCAGAGGCAGGGAGGGGAAAGGAGGGAGACTGGAATGAAAAGAAAAGGAAATGAGCAAAGAAAACCATAATAGACTGATAGAGAGAAAGCCAGAAGTCACCTCTGGGTGGGTAGGGGGTTGGAAGAGGGGAAATAGCCTCCCGCACTAGGAATGCCCTTTATTTGGATCCAGATGATAAGTTGATAGTCATTAAAGCTCATTGAGCACTTAAAATCCATGCACTTTGCTGTATTTTATTTGACCTTAGTTAAAATAAAACCAGAAAACTGTG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000032292-Nr2e3:NM_013708:7
Average complexity
IR-S
Mappability confidence:
NA
Protein Impact
Alternative protein isoforms
No structure available
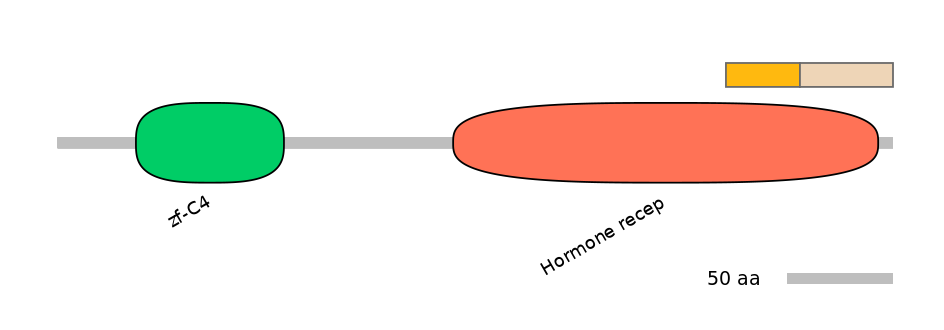
Features
Disorder rate (Iupred):
C1=0.194 A=NA C2=0.000
Domain overlap (PFAM):
C1:
PF0010425=Hormone_recep=FE(17.3=100)
A:
NA
C2:
PF0010425=Hormone_recep=PD(18.3=82.2)
Main Inclusion Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Rat
(rn6)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
No suggested primer sequences
R:
No suggested primer sequences
Band lengths:
Functional annotations
There are 1 annotated functions for this event
PMID: 33007388
NR2E3 produces a large isoform encoded in 8 exons and a shorter isoform of 7 exons. The short isoform may have a physhiological role, since intron 7 retention (MmuINT0110513, but normally as APA) for the generation of the short isoform appears to be developmentally regulated and has been identified in transcriptomic analysis at least in humans and mice. The in-frame stop codon at the very beginning of intron 7 is also evolutionary conserved in ten of the eleven vertebrate species considered, pointing to some functionality. This short isoform displays the N-terminal transactivator domain, can bind their target motifs and potentially interact with CRX through the DBD, but would lack the C-terminal domains H10 and AF2 (respectively involved in the dimerization and repression function of NR2E3), which are also highly conserved across species. Note: the short isoform is normally produced by APA, but standard IR would generate the equivalent protein.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types
Other AS DBs: