RnoALTA0018302-5/5 @ rn6
Alternative 3'ss
Gene
ENSRNOG00000036839 | Hnrnpa1
Description
heterogeneous nuclear ribonucleoprotein A1 [Source:RGD Symbol;Acc:69234]
Coordinates
chr7:144867633-144868708:+
Coord C1 exon
chr7:144867633-144867707
Coord A exon
chr7:144868574-144868672
Coord C2 exon
chr7:144868673-144868708
Length
99 bp
Sequences
Splice sites
5' ss Seq
ATGGTAAGT
5' ss Score
11.01
3' ss Seq
TGGCTTGTCAGTCATCAAAGACT
3' ss Score
-4.98
Exon sequences
Seq C1 exon
GTGGCTTTGGTGGCAGCCGTGGTGGTGGTGGATATGGTGGCAGTGGGGATGGCTATAATGGATTTGGCAATGATG
Seq A exon
ACTTTTCCATTTTAGGAAGCAATTTTGGAGGTGGTGGAAGCTACAATGATTTTGGCAATTACAACAACCAGTCATCAAATTTTGGACCGATGAAAGGAG
Seq C2 exon
GAAACTTTGGAGGCAGGAGCTCTGGCCCTTATGGTG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000036839-15-20,15-19,15-18,15-17,15-15-5/5
Average complexity
Alt3
Mappability confidence:
NA
Protein Impact
Protein isoform when splice site is used (No Ref, Alt. Stop)
No structure available
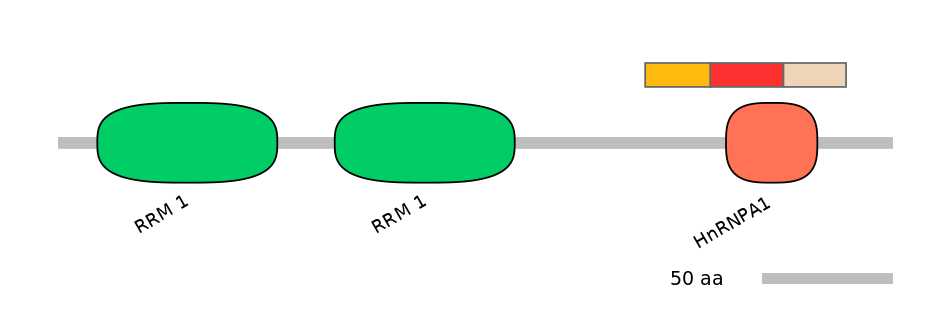
Features
Disorder rate (Iupred):
C1=0.635 A=NA C2=0.870
Domain overlap (PFAM):
C1:
PF116273=HnRNPA1=PU(2.8=3.8)
A:
NA
C2:
PF116273=HnRNPA1=PD(94.4=73.9)
Main Inclusion Isoform:
NA
Other Inclusion Isoforms:
NA
Associated events
Conservation
Human
(hg19)
No conservation detected
Mouse
(mm9)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CTTTGGTGGCAGCCGTGG
R:
CCATAAGGGCCAGAGCTCCT
Band lengths:
105-204
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]