RnoALTA0041419-4/4 @ rn6
Alternative 3'ss
Gene
ENSRNOG00000000184 | Tmprss6
Description
transmembrane serine protease 6 [Source:RGD Symbol;Acc:1307138]
Coordinates
chr7:119663671-119664627:-
Coord C1 exon
chr7:119664514-119664627
Coord A exon
chr7:119663759-119663845
Coord C2 exon
chr7:119663671-119663758
Length
87 bp
Sequences
Splice sites
5' ss Seq
AAGGTAGAG
5' ss Score
6.38
3' ss Seq
TCTGGGTCTCACTTTGGAAGGCT
3' ss Score
3.5
Exon sequences
Seq C1 exon
TCTGCAGAGCCATGTTCCAGTGCCAAGAGGACAGCACGTGCATCTCACTGCCGAGAGTCTGTGACCGGCAGCCCGACTGTCTCAATGGTAGCGACGAAGAGCAGTGCCAAGAAG
Seq A exon
GCTCCCTGGTTGCTCAGAGACCCACCTTCAGTGGCCTGCCCGGTCCCCTCCATCCCAGGAGTGCCCTGTGGGACATTCACTTTCCAG
Seq C2 exon
TGTGAGGACCGGAGCTGTGTGAAGAAGCCCAACCCCGAGTGTGACGGGCAGGCAGACTGCAGGGATGGCTCGGATGAGGAGCACTGTG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000000184-17-21,17-20,17-19,17-18-4/4
Average complexity
Alt3
Mappability confidence:
NA
Protein Impact
ORF disruption when splice site is used (sequence inclusion)
No structure available
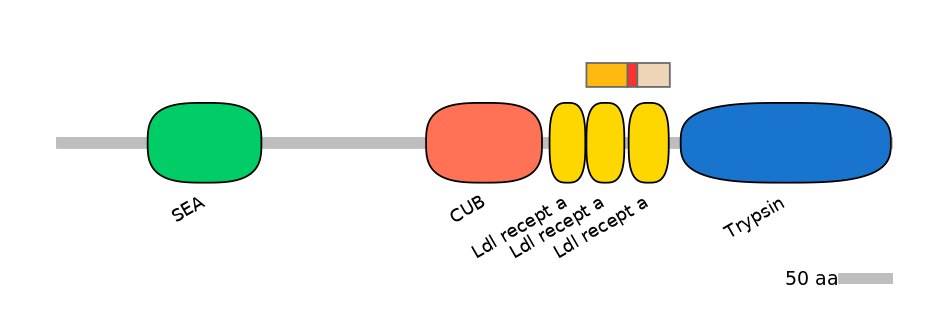
Features
Disorder rate (Iupred):
C1=0.000 A=NA C2=0.000
Domain overlap (PFAM):
C1:
PF0005713=Ldl_recept_a=WD(100=92.3)
A:
NA
C2:
PF0005713=Ldl_recept_a=WD(100=95.0)
Main Inclusion Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Mouse
(mm9)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GTTCCAGTGCCAAGAGGACAG
R:
CTCCTCATCCGAGCCATCCC
Band lengths:
182-269
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]