RnoEX0073037 @ rn6
Exon Skipping
Gene
ENSRNOG00000050210 | RT1-CE10
Description
RT1 class I, locus CE10 [Source:RGD Symbol;Acc:1359292]
Coordinates
chr20:154897-155614:-
Coord C1 exon
chr20:155339-155614
Coord A exon
chr20:155107-155244
Coord C2 exon
chr20:154897-154929
Length
138 bp
Sequences
Splice sites
3' ss Seq
CCTGTACCTGTCCTTCCCAGAGC
3' ss Score
8.66
5' ss Seq
CAGGTAGGA
5' ss Score
9.79
Exon sequences
Seq C1 exon
ATCCCCCAGAGGCACATGTGACCCTTCACCCCAGACCTGAAGGTGATGTCACCCTGAGGTGCTGGGCCCTGGGCTTCTACCCTGCTGACATCACATTGACCTGGCAGTTGAATGGGGAGGACCTAACCCAGGACATGGAGTTTGTGGAGACCAGGCCTGCAGGGGATGGAACCTTCCAGAAGTGGGCATCTGTGGTGGTGCCTCTTGGGAAGGAGCAGAATTACACATGCCTTGTGGAGCATGAGGGGCTGCCTGAGCCTCTTACCCAGAGATGGG
Seq A exon
AGCCTTCTCCATCCACCAACACCAACATGGGAATCGATGTTACTTCTGTTTTTCTTGGACCTTTGGCTGTCTTTGTAGTTTTGGCCATCATTGGAGCTGCGGTGGCTGTTGTGAGGAGGAGGAGGAGGAGAAACACAG
Seq C2 exon
GTGGGGAAGGAGGAGACTACACCCCTGCTCCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000050210-'5-7,'5-4,6-7
Average complexity
C1
Mappability confidence:
78%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.265 A=0.291 C2=1.000
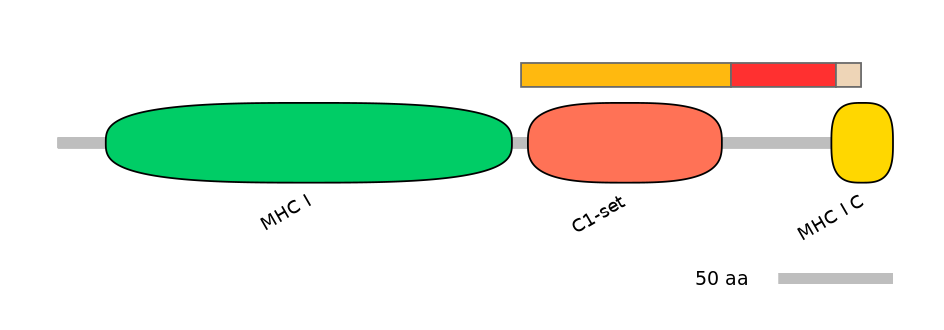
Domain overlap (PFAM):
C1:
PF0765410=C1-set=WD(100=92.5)
A:
PF066236=MHC_I_C=PU(14.3=4.3)
C2:
PF066236=MHC_I_C=FE(39.3=100)
Main Skipping Isoform:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg19)
No conservation detected
Mouse
(mm9)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CTGAGGTGCTGGGCCCTG
R:
GGTGTAGTCTCCTCCTTCCCC
Band lengths:
246-384
Functional annotations
There are 3 annotated functions for this event
PMID: 2441874
Qa region gene Q7 encodes a membrane-bound as well as a secreted form of the serologically defined antigen Qa-2. The Q7 gene introduced into liver-derived cells is expressed as a membrane-bound and as a secreted molecule. In transfected L cells it is expressed only as a soluble protein. Biochemical analysis suggests that the Q7 product is anchored to the liver cell membranes by a phospholipid tail.
PMID: 3486423
Polyclonal activation of T lymphocytes induces a 3- to 4-fold increase in the biosynthesis of Qa-2 molecules but no increase in cell-surface levels. Analysis of the biosynthetic pathway of the Qa-2 molecule in activated lymphocytes reveals that approximately equal to 70% of the newly synthesized Qa-2 molecules are secreted as soluble molecules. In resting-cell populations, Qa-2 remains entirely cell-associated. This process is unique to the Qa-2 molecule, since other class I molecules (e.g., H-2Kb and H-2Db) synthesized by activated cells remain cell-associated.
PMID: 8127900
AS event changes a membrane bound form into a soluble form. Critical to these experiments was preparation of an anti-peptide antiserum against an epitope encoded by a junction of exon 4 and exon 6. Supernatants of splenocytes cultured in vitro as well as serum or plasma contain two forms of soluble Qa-2 molecules. One form corresponds to a secreted molecule translated from transcripts from which exon 5 has been deleted; the other is derived from membrane-bound antigens or their precursors. The levels of both soluble forms of Qa-2 are inducible upon stimulation of the immune system, suggesting an immunoregulatory role for these molecules or for the mechanism leading to the reduction of cell-associated Qa-2 antigens in vivo.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]