RnoEX6015309 @ rn6
Exon Skipping
Gene
ENSRNOG00000056219 | Olr1
Description
oxidized low density lipoprotein receptor 1 [Source:RGD Symbol;Acc:620515]
Coordinates
chr4:163239849-163244241:-
Coord C1 exon
chr4:163244105-163244241
Coord A exon
chr4:163242838-163242953
Coord C2 exon
chr4:163239849-163242562
Length
116 bp
Sequences
Splice sites
3' ss Seq
AGAGTGTTGTCTTTCCACAGAAC
3' ss Score
8.75
5' ss Seq
ATTGTAAGT
5' ss Score
8.54
Exon sequences
Seq C1 exon
GTCCTTGTCCACAAGACTGGATCTGGCATAAAGAAAACTGTTACCTCTTCCATGGGCCCTTTAACTGGGAAAAAAGTCGGGAGAATTGCCTATCTTTAGATGCCCAGTTACTACAAATTAGTACCACAGATGATCTG
Seq A exon
AACTTCGTCTTACAAGCAACTTCCCATTCCACCTCCCCATTTTGGATGGGATTACATCGGAAAAATCCCAACCACCCATGGCTATGGGAGAACGGCTCTCCTTTGAGTTTTCAATT
Seq C2 exon
CTTTAGGACCAGGGGCGTTTCTTTACAGATGTACTCATCAGGCACCTGTGCATATATTCAAGGAGGAGTTGTGTTTGCTGAAAACTGCATTTTAACTGCATTCAGCATATGTCAGAAGAAGGCAAATTTATTGCTAACTCAGTGAAACTAAGGATTCTGGAGAAGAACAGGAGAAGACCTTTAACTGTTGTTTTGAAATTTAAGCTATCCTTTCTTGGGTGTAAAACATGTGGCCTTGACAGCTGTCAGTTACTTTCTAACTGCAGTTCACCTCAACAGAGACAAAGACCAGAAGCAAAAACCCGGGGGTCCAGCTGATGGCATCTTTGTATCAAAAGTTGTGAATTCAATTGTTTATCCATGTACACTGGCCCCGCCCCTCCCAAGACTCCCAACCAACCTGCAATCCTTTTTTTCTTTCTTGTTTTAAACTATGCCTCCTGTCTGACCTGGGGGATGCTTTCTGCTCAATTTCCTCTACCTCAGGTATGCCTTCTGTT
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000056219-'8-9,'8-8,9-9=AN
Average complexity
A_S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref, Alt. Stop)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
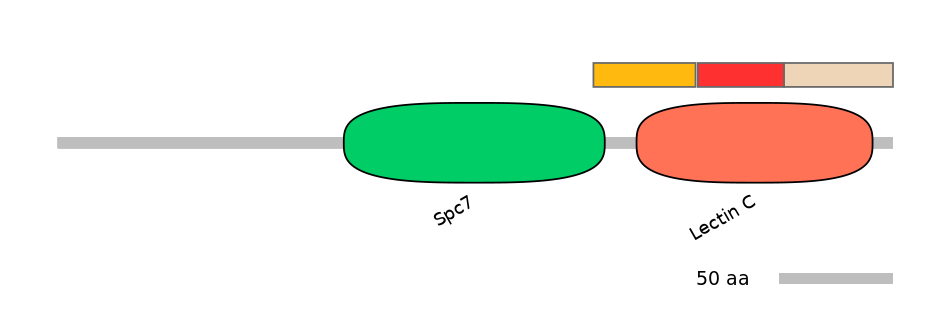
Domain overlap (PFAM):
C1:
PF083176=Spc7=PD(4.3=10.9),PF0005916=Lectin_C=PU(24.8=56.5)
A:
PF0005916=Lectin_C=FE(36.2=100)
C2:
PF0005916=Lectin_C=PD(37.1=79.6)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg19)
No conservation detected
Mouse
(mm9)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TCCACAAGACTGGATCTGGCA
R:
TGCCTTCTTCTGACATATGCTGA
Band lengths:
254-370
Functional annotations
There are 2 annotated functions for this event
PMID: 15976314
Macrophages from subjects carrying the non-risk disease haplotype at OLR1 gene have an increased expression of LOXIN (skipping exon 5) at mRNA and protein level, which results in a significant reduction of apoptosis in response to oxLDL. Expression of LOXIN in different cell types results in loss of surface staining, indicating that truncation of the C-terminal portion of the protein has a profound effect on its cellular trafficking. Furthermore, the proapoptotic effect of LOX-1 receptor in cell culture is specifically rescued by the coexpression of LOXIN in a dose-dependent manner.
PMID: 18191942
LOXIN lacks part of the C-terminus lectin-like domain. In vivo and in vitro studies support that the new splicing isoform is protective against acute myocardial infarction. The mechanism by which LOXIN exerts its protective role is unknown. In this paper the authors report studies on the heterologous expression and functional characterization of LOXIN variant in mammalian fibroblasts and human endothelial cells. The authors found that LOXIN, when expressed in the absence of LOX-1, shows diminished plasma membrane localization and is deficient in ox-LDL ligand binding. When co-transfected with the full-length counterpart LOX-1, the two isoforms interact to form LOX-1 oligomers and their interaction leads to a decrease in the appearance of LOX-1 receptors in the plasma membrane and a marked impairment of ox-LDL binding and uptake. Co-immunoprecipitation studies confirmed the molecular LOX-1/LOXIN interaction and the formation of non-functional hetero-oligomers.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]