BtaALTA0023614-2/2 @ bosTau6
Alternative 3'ss
Gene
ENSBTAG00000004400 | PRRC2B
Description
proline-rich coiled-coil 2B [Source:HGNC Symbol;Acc:HGNC:28121]
Coordinates
chr11:101611657-101616153:+
Coord C1 exon
chr11:101611657-101611849
Coord A exon
chr11:101615835-101615837
Coord C2 exon
chr11:101615838-101616153
Length
3 bp
Sequences
Splice sites
5' ss Seq
CAGGTACCG
5' ss Score
10.05
3' ss Seq
CTCTGACCGTCTTCTCTCAGCAG
3' ss Score
6.01
Exon sequences
Seq C1 exon
GAACAGTTGGGACCCCAGGAGGCAGCGGCAGTTATCCTTGAGCTCTGCAGACAGTGCTGATGCCAAGCGCACCCAAGAGGAAGGAAAAGACTGGAGTGAAACAGGAGGCGTGACCCGTGTCGTCCGGAAGGTGCCGGAACCTCAGCCGCCTTCCAGGAAGCTGCACAGCTGGGCGTCAGGCCCTGACTACCAG
Seq A exon
CAG
Seq C2 exon
AAATCCTCCATGGGCACTGTATTTCGGCAGCAGTCTGCCGAGGACAAAGAGGACAAGCCCCCACCGCGGCAGAAGTTTGTCCAGTCTGAGATGTCCGAGGCTGTGGAGCGAGCCCGGAAGCGCCGGGAGGAGGAGGAGCGCCGTGCGCGCGAGGAAAGGCTGGCTGCCTGTGCTGCCAAACTCAAGCAGCTTGACCAGAAGTGCAAGCAGGCTCAGCGGGCCAGCGAGGCCCAGAAGCAGGCGGAGAAGGAAGCTCCCCGCTCTCCGGGGGCTGAGAAGGTGACTCCCCAGGAGAACGGCCCTGCCGTCCACAAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSBTAG00000004400-13-22,13-21-2/2
Average complexity
Alt3
Mappability confidence:
NA
Protein Impact
Protein isoform when splice site is used (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=1.000 A=NA C2=1.000
Domain overlap (PFAM):
C1:
NO
A:
NA
C2:
NO
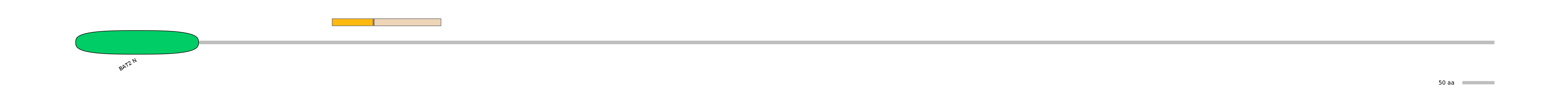
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Mouse
(mm10)
No conservation detected
Rat
(rn6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CCTTCCAGGAAGCTGCACAG
R:
CTTGTCCTCTTTGTCCTCGGC
Band lengths:
102-105
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development