BtaEX0000199 @ bosTau6
Exon Skipping
Description
NA
Coordinates
chr16:77484306-77491642:-
Coord C1 exon
chr16:77491582-77491642
Coord A exon
chr16:77488021-77488109
Coord C2 exon
chr16:77484306-77485524
Length
89 bp
Sequences
Splice sites
3' ss Seq
TTTCCCCTTCCTCTGTTCAGCAT
3' ss Score
9.66
5' ss Seq
AAGGTATCT
5' ss Score
7.56
Exon sequences
Seq C1 exon
TTATTGGCGTTGGATTATTGCTCTGCCTGTACTGCTGTTTTTGCAGACAGAGGAAGAAAGG
Seq A exon
CATATACGTAACTGGTGAGAGCCACAGACAAGACATTTTATTTCTCTCTGAGAAGAGATGAGAGAAACGTTTGCTTTTATCATTAAAAG
Seq C2 exon
GAAAGCAGAATGTAGCGCTACGTACACCACTTATCAGGATAAAGCAACCACTGCAACAGAACAGATGAACTGAACCGTTTCCAGGAACGCATGTACCATTCAGCCATGAAAATGAAGGAAACCTTGCCATTTGCCACAATACAGACAGACCTGAGGGGACTTTGCTGGGTAATAAGTCATGTAAAGAAAGACGTGTACTGTCTGATCTCACTTACACATGGAATCGAAAAAAACAAGCTTTCACAGAAGAAAAATGAGATTTGTCGATGTCAGCGGTGGGGGACTGGTGGGGGTGGGTTTGAGTAAAGGTGGTCAGAAAATACAAACCTTCAGTTATAGGGTGAGAAAGTACTGGGGTATAATTACAGCACAGTGGCTACCATGAGTTACTCATACTGTATTTATGTTGGCAAGTTGCTAAGAGAGTGGATCTTAAAAAGTTCTAATCACATGGAAGAAAAAAGCTTTTTGTAGTATTGACATTATCAAATCATGCTGTA
VastDB Features
Vast-tools module Information
Secondary ID
ENSBTAG00000005397_MULTIEX1-13/13=11-C2
Average complexity
C1
Mappability confidence:
86%=86=100%
Protein Impact
In the CDS, with uncertain impact
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.208
Domain overlap (PFAM):
C1:
PF0248011=Herpes_gE=FE(24.7=100),PF041569=IncA=FE(33.3=100)
A:
NO
C2:
PF0248011=Herpes_gE=PD(24.7=80.0),PF041569=IncA=PD(36.7=88.0)
Main Inclusion Isoform:
ENSBTAT00000007107fB42
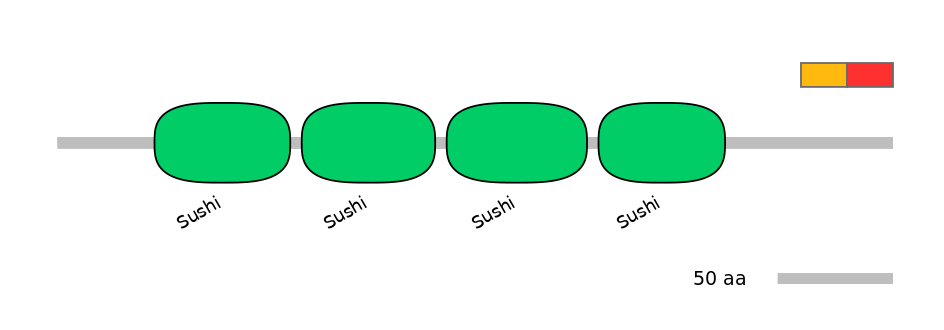
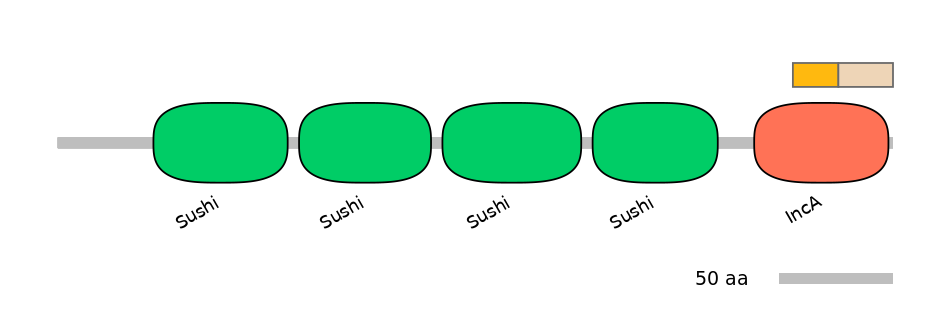
Other Skipping Isoforms:
NA
Associated events
Conservation
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GCTCTGCCTGTACTGCTGTTT
R:
GGCAAATGGCAAGGTTTCCTT
Band lengths:
177-266
Functional annotations
There are 1 annotated functions for this event
PMID: 33767911
The skipping is unregulated in bladder cancer. In BCa cell lines, enforced expression of CD46-CYT2 (exon 13-skipping isoform) promoted, and CD46-CYT1 (exon 13-containing isoform) attenuated,cell growth, migration, and tumorigenicity in a xenograft model. Heterogeneous nuclear ribonucleoprotein (hnRNP)A1 is a novel CYT2 binding partner, and this interaction facilitates the interaction of hnRNPA1 with IRES RNA to promote IRES-dependent translation of HIF1a and c-Myc.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development