BtaEX6071815 @ bosTau6
Exon Skipping
Gene
ENSBTAG00000040336 | ZNF268
Description
Bos taurus zinc finger protein 268 (ZNF268), mRNA. [Source:RefSeq mRNA;Acc:NM_001083646]
Coordinates
chr17:45191784-45217459:+
Coord C1 exon
chr17:45191784-45191839
Coord A exon
chr17:45209365-45209556
Coord C2 exon
chr17:45217333-45217459
Length
192 bp
Sequences
Splice sites
3' ss Seq
GCTTGCTCTTGGGTCTACAGGTG
3' ss Score
8.41
5' ss Seq
TGGGTGAGT
5' ss Score
8.73
Exon sequences
Seq C1 exon
GGAACTCAAAACTGGCTGTGGGGATGGCCACCAGGGTCCGGACAGCTGCCATCTGG
Seq A exon
GTGCCACCTCTCCAGGAGCGAGACAGTTCATGCAATGTGAGCAGAAAGCTCCAAAGTGAGAAATCCATCCTGGATCAAGGGACCCCTGACCAGAAGCCTCTCTCTGGAGGACCCTGGCAGAGGCCCAGGAGACCTGGGGTCCTGGAGTGGCTGCTTATTTCCCAGGACCAGCCGAAAGCCAAGAAATCCTGG
Seq C2 exon
GGACCATTGTCATTCATGGATGTGTTTGTGGACTTTACCTGGGAAGAGTGGCAGCTGCTGGACTCAGCTCAGAAGCACCTGTACAGGAGTGTGATGTTGGAGAACTTTAGAAACCTGGTGTCCCTGG
VastDB Features
Vast-tools module Information
Secondary ID
ENSBTAG00000040336-'3-9,'3-7,7-9=AN
Average complexity
A_C1
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.312 C2=0.000
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
PF0135222=KRAB=WD(100=95.3)
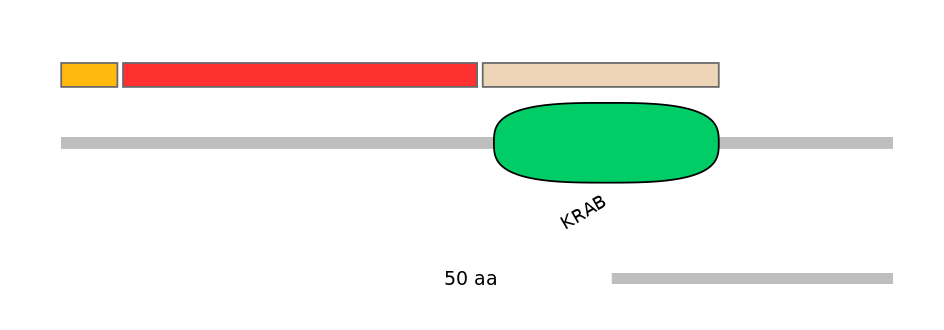
Main Skipping Isoform:
ENSBTAT00000056095fB7983
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
AACTCAAAACTGGCTGTGGGG
R:
CCAGGGACACCAGGTTTCTAA
Band lengths:
181-373
Functional annotations
There are 1 annotated functions for this event
PMID: 16865230
mRNA: ZNF268a (skipping exon 6), ZNF268b (all in), ZNF268c (4,6), ZNF268d (3,4,6), ZNF268e (4,5,6), ZNF268f (3,4), ZNF268g (3). Subcellular localization analysis showed that ZNF268a and ZNF268b2 distributed diffusely throughout the cell, while ZNF268b1 mainly localized in the cytoplasm. Moreover, using a CAT reporter system fused to the Gal4 DNA binding domain of the ZNF268 gene, the ZNF268a and b2 activated the CAT reporter gene expression, while the KRAB domain, corresponding to the ZNF268b1 repressed the reporter gene expression. mRNA: ZNF268a (skipping exon 6 = HsaEX0072991), ZNF268b (all in), ZNF268c (4,6), ZNF268d (3,4,6), ZNF268e (4,5,6), ZNF268f (3,4), ZNF268g (3). Protein: ZNF268a, ZNF268b1 (PTC in last exon), ZNF268b2 (from internal ATG in exon 7, derived from isoforms c-g). Exon 3 =HsaEX0072986, Exon 4 = HsaEX0072989, Exon 5 = HsaEX0072990, Exon 6 = HsaEX0072991.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]