DmeALTA0000086-2/2 @ dm6
Alternative 3'ss
Gene
FBgn0000015 | Abd-B
Description
The gene Abdominal B is referred to in FlyBase by the symbol DmelAbd-B (CG11648, FBgn0000015). It is a protein_coding_gene from Dmel. It has 11 annotated transcripts and 11 polypeptides (4 unique). Gene sequence location is 3R:16927212..16972236. Its molecular function is described by: DNA-binding transcription factor activity; RNA polymerase II cis-regulatory region sequence-specific DNA binding; transcription regulatory region sequence-specific DNA binding. It is involved in the biological process described with 25 unique terms, many of which group under: insemination; spiracle morphogenesis, open tracheal system; genital disc anterior/posterior pattern formation; positive regulation of transcription, DNA-templated; gland development. 237 alleles are reported. The phenotypes of these alleles manifest in: imaginal tissue; presumptive embryonic/larval system; non-connected developing system; male accessory gland secondary cell; imaginal disc primordium. The phenotypic classes of alleles include: cell number defective; sterile; increased mortality; phenotype. Summary of modENCODE Temporal Expression Profile: Temporal profile ranges from a peak of moderate expression to a trough of very low expression. Peak expression observed at stages throughout embryogenesis.
Coordinates
chr3R:16931116-16931613:-
Coord C1 exon
chr3R:16931402-16931613
Coord A exon
chr3R:16931315-16931317
Coord C2 exon
chr3R:16931116-16931314
Length
3 bp
Sequences
Splice sites
5' ss Seq
GGTGTAAGT
5' ss Score
8.07
3' ss Seq
ACCTCATTTCTCCCCCGCAGCAG
3' ss Score
10.52
Exon sequences
Seq C1 exon
ACATCAATATGTCCATACAATTAGCGCCACTGCATATACCCGCCATCCGGGCCGGTCCGGGATTCGAGACGGACACCTCGGCGGCGGTCAAGCGGCACACGGCACACTGGGCCTACAACGACGAGGGATTCAATCAGCATTACGGCTCCGGGTACTACGACCGCAAGCACATGTTCGCCTATCCTTACCCAGAAACGCAGTTTCCGGTTGGT
Seq A exon
CAG
Seq C2 exon
TACTGGGGCCCCAACTACCGCCCCGATCAGACCACCTCTGCCGCAGCGGCGGCGGCCTACATGAACGAAGCGGAGCGCCACGTGAGCGCCGCCGCCCGACAGTCCGTCGAGGGCACATCGACGTCCAGCTATGAGCCGCCCACCTACTCCTCGCCAGGCGGCCTGCGCGGCTATCCCAGCGAGAACTACTCCAGCTCAG
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0000015-10-8,10-7-2/2
Average complexity
Alt3
Mappability confidence:
NA
Protein Impact
Protein isoform when splice site is used (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.163 A=NA C2=0.729
Domain overlap (PFAM):
C1:
NO
A:
NA
C2:
NO
Main Inclusion Isoform:
FBpp0082826
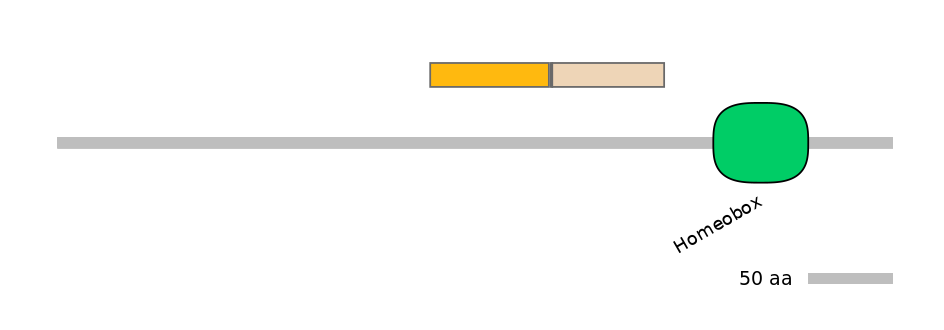
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
FBpp0082823, FBpp0082824, FBpp0082825, FBpp0089276, FBpp0303552, FBpp0312359
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GTACTACGACCGCAAGCACAT
R:
TCTGATCGGGGCGGTAGTTG
Band lengths:
92-95
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)