DmeALTA0009653-1/2 @ dm6
Alternative 3'ss
Gene
FBgn0010328 | woc
Description
The gene without children is referred to in FlyBase by the symbol Dmelwoc (CG5965, FBgn0010328). It is a protein_coding_gene from Dmel. It has 6 annotated transcripts and 6 polypeptides (5 unique). Gene sequence location is 3R:27256595..27264055. Its molecular function is described by: zinc ion binding; chromatin binding; protein binding; DNA-binding transcription factor activity, RNA polymerase II-specific. It is involved in the biological process described with: ecdysone biosynthetic process; regulation of transcription, DNA-templated; regulation of transcription by RNA polymerase II. 30 alleles are reported. The phenotypes of these alleles manifest in: circulatory system; cellular anatomical entity; organism subdivision; intracellular non-membrane-bounded organelle; intracellular organelle. The phenotypic classes of alleles include: lethal; lethal - all die before end of pupal stage; phenotype; increased mortality during development. Summary of modENCODE Temporal Expression Profile: Temporal profile ranges from a peak of very high expression to a trough of moderate expression. Peak expression observed within 00-06 hour embryonic stages.
Coordinates
chr3R:27260091-27262487:-
Coord C1 exon
chr3R:27262227-27262487
Coord A exon
NA
Coord C2 exon
chr3R:27260091-27261940
Length
0 bp
Sequences
Splice sites
5' ss Seq
CTGGTAAGT
5' ss Score
10.65
3' ss Seq
TCCCCTTTTTTTGCCAGCAGATG
3' ss Score
8.76
Exon sequences
Seq C1 exon
ACACAGTGGAAAATGTTTTGGAGCCAGAGGAATCAGATGTTTGCCTTATTCCCGATGATCAAGAAACGGAAGTGACTGAAGCTGAGAAGGAACTGGCCCGCGAGAATGCCGAGAAGGCCGCCGAAGAGGAGGAAGCCGAGGAGCGGCAAGCCAAGACACCCGATAACGATAGAAAGTCCCCGGCAGATGCCGCCGCCGATCCAGAAGACGACAGTGCAGCTGCCGTTGACGCCACCGACGTATCTCCAACCACGCCAACTG
Seq A exon
NA
Seq C2 exon
ATGGAGTAGCTGCAGCAGCTAACGAAGAGGAGGCCGGTGGCGATGTTCCCGATCCCGACGAGCCCACAGCCGAGGAATCCTCCAGTGCCGGAGCGGGTGAACCTGGTTCCACGCCGAAAATCATCATTAGTTGGATCGTTGGCAGCGTTCAGAAGTGCAAACAGTGTGGCGATGAAAAGAATTGTGGGTTTCGGTACAGGAGTCCAGAGGAGTCAGAAGAAATGGATAAGGAGAAGGAAAACGATAAGGCAGAGGAGAACGCTGAGGAGGGGGAGGAGGAGGAGGCTCGGCAAAAGCCTGCTGCTCCCAGTGGGTTCGAGTACATCTGCGATACCGCCTGTGTCGACGCCCTGCTAGAAGATCAACCCGGCAAGTACTTTGTACGCCGCAAGAAGTTCCTGGTCGAGGAAGTGGTTCGAGAAGAAGGCGCCGCCGAAGCGGAGGCTAGCGCCGATGGCGAGGGCGGAGATGCCGAGGAGGAAGCTACTCCAGCCAACACG
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0010328-5-5,5-4-1/2
Average complexity
Alt3
Mappability confidence:
NA
Protein Impact
Protein isoform when splice site is used (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=1.000 A=NA C2=0.281
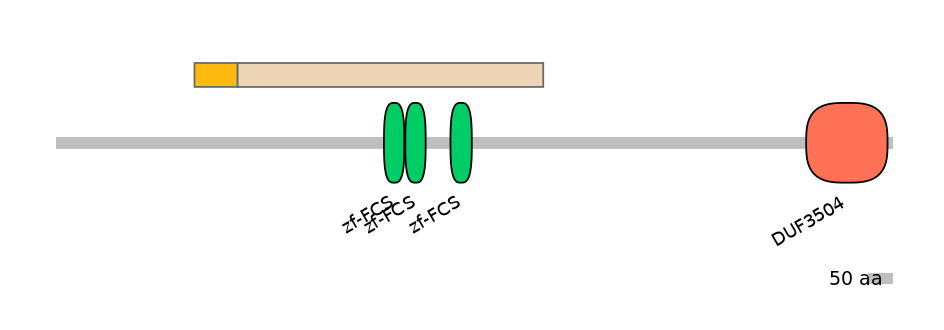
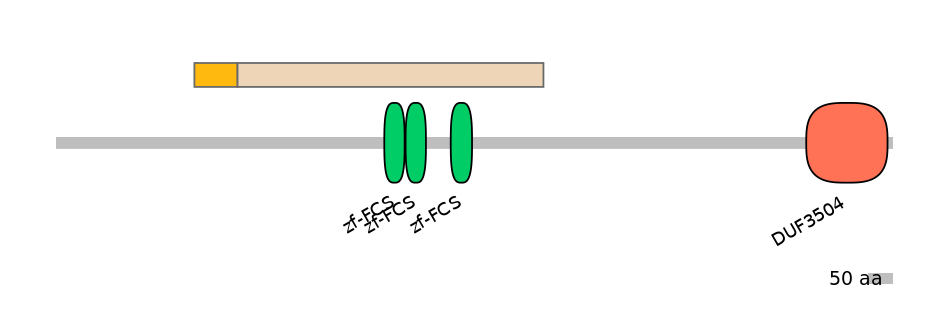
Domain overlap (PFAM):
C1:
NO
A:
NA
C2:
PF064679=zf-FCS=WD(100=6.8),PF064679=zf-FCS=WD(100=6.8),PF064679=zf-FCS=WD(100=7.1)
Main Inclusion Isoform:
FBpp0306778
Main Skipping Isoform:
FBpp0084548
Other Inclusion Isoforms:
FBpp0306779
Other Skipping Isoforms:
FBpp0111770
Associated events
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CGATCCAGAAGACGACAGTGC
R:
CTCCTCTTCGTTAGCTGCTGC
Band lengths:
97-100
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)