DmeEX0004823 @ dm6
Exon Skipping
Gene
FBgn0265042 | Irk1
Description
The gene Inwardly rectifying potassium channel 1 is referred to in FlyBase by the symbol DmelIrk1 (CG44159, FBgn0265042). It is a protein_coding_gene from Dmel. It has 8 annotated transcripts and 8 polypeptides (5 unique). Gene sequence location is 3R:23168317..23185642. Its molecular function is described by: inward rectifier potassium channel activity. It is involved in the biological process described with 6 unique terms, many of which group under: transport; regulation of membrane potential; inorganic ion transmembrane transport; developmental process; locomotory behavior. 19 alleles are reported. The phenotypes of these alleles manifest in: trichogen cell; wing; mesothoracic tergum; wing vein. The phenotypic classes of alleles include: lethal - all die during P-stage; visible; locomotor rhythm defective; viable; some die during pupal stage; partially lethal - majority die.
Coordinates
chr3R:23173873-23184164:+
Coord C1 exon
chr3R:23173873-23174818
Coord A exon
chr3R:23182946-23183060
Coord C2 exon
chr3R:23183444-23184164
Length
115 bp
Sequences
Splice sites
3' ss Seq
ATCTTGTTGTTCCGTTTCAGGTA
3' ss Score
11.59
5' ss Seq
CAGGTGAGC
5' ss Score
9.6
Exon sequences
Seq C1 exon
TGCCTCACACACGAGTTTATCCAACGATAACGTCATCCGCTTTGCCAACAGTATTAAAGTTAATCTGAAAAGTGCTCGAATCTTTTGTTTTCAATCGTCATAATTGTAATAGCAACAACTACAAAATCAAATTATAAATAATAATACCAACAACAATTGTAGCCAAGGTTGCCGCGAACAGTTTCGTTTCGTTGTTGTTTTGCCCGGTGAAGGAGGAGCTGTCAATTCAACATCTCTGCCGACGTCGCTGTTGCCGTTACATTTCAGTTGCTGCCATCGCATAAAAACCAAACAGAGATCCCCAAACCGCGACAGAAACAGCAAATTTACATAGCATTATACACGTTACACAACGCAGCAGCGCGTGCCAGTGCAAAATCCCAAAAACACCTAAGCCAAATCGAAATCCTCGAGTCCATACAAAACACCACGAAATTCCTTTTGGCCAAAATGAAGCGTCTGATGACCTGGGAGCGCGATCTGGTGGATGCCATGTACGA
Seq A exon
GTACCGGCAGACACGTTTCAGTTCGCGGCGTGTTCGAAAACGTGTGGTCTTCAAGCACGGCGAGTGCAACGTTGTGCAGGGAAATGTGGCCAAGAGACGGCGTCGCTATCTGCAG
Seq C2 exon
GACATCTTCACCACCCTGGTTGACGCCCAGTGGCGCTGGACGCTGCTCGTCTTCGCCGCCAGCTTCGTGTTCTCGTGGGCCTTCTTTGGATTCATCTGGTGGATCATCGCCTACGCACACAATGATCTGGAGTACACCAATCTCAAGAACCAGTCACCCGATCTGGTGGCCAACATAACGCACACGGTCTGCGTAACACAGGTCTCCAATATGATGTCCGCCTTCCTGTACTCCGTGGAAACCCAGACGACGATAGGCTATGGTAATCGCTATGTGACGGAGGAGTGCCCGGAGGCGATATTCACGATGTGCATCCAGTGCATCACGGGTGTCTTCATCCAGGCGTTCATGGTGGGCATTGTGTTTGCCAAGTTGTCGCGTCCCAAGAAACGTGCCCAGACGCTGCTCTTCTCGCGCAATGCTGTTATCTGCCACCGGGATGGAGTTCCATGCCTGATGTTCCGTGTGGGAGACATGCGCAAGTCGCACATCATCGAGGC
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0265042-'5-11,'5-10,11-11
Average complexity
C2
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
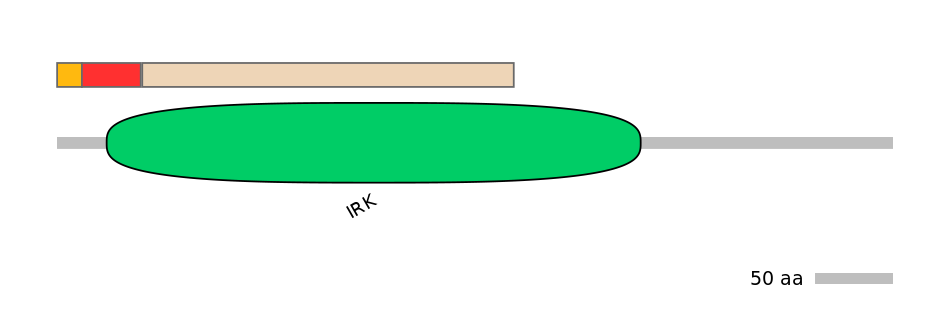
Domain overlap (PFAM):
C1:
NO
A:
PF0100715=IRK=PU(6.4=56.4)
C2:
PF0100715=IRK=FE(69.4=100)
Main Inclusion Isoform:
FBpp0307577
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
FBpp0307576, FBpp0307578, FBpp0307579, FBpp0307580, FBpp0307581, FBpp0307582
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TCTGGTGGATGCCATGTACGA
R:
AGTACAGGAAGGCGGACATCA
Band lengths:
253-368
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)