DmeEX0005111 @ dm6
Exon Skipping
Gene
FBgn0264695 | Mhc
Description
The gene Myosin heavy chain is referred to in FlyBase by the symbol DmelMhc (CG17927, FBgn0264695). It is a protein_coding_gene from Dmel. It has 22 annotated transcripts and 22 polypeptides (20 unique). Gene sequence location is 2L:16766566..16788766. Its molecular function is described by 6 unique terms, many of which group under: nucleoside-triphosphatase activity; catalytic activity; hydrolase activity, acting on acid anhydrides; drug binding; actin filament binding. It is involved in the biological process described with 15 unique terms, many of which group under: epithelial cell development; adult somatic muscle development; regulation of biological quality; reproductive process; protein stabilization. 161 alleles are reported. The phenotypes of these alleles manifest in: somatic cell of ovariole; muscle attachment site; embryonic/larval heart; adult visceral muscle; germline cell. The phenotypic classes of alleles include: sterile; fertile; phenotype; increased mortality during development.
Coordinates
chr2L:16772437-16774857:+
Coord C1 exon
chr2L:16772437-16772540
Coord A exon
chr2L:16773277-16773380
Coord C2 exon
chr2L:16774450-16774857
Length
104 bp
Sequences
Splice sites
3' ss Seq
TTCCAACTATGTGTATGTAGAGA
3' ss Score
4.19
5' ss Seq
GATGTAAGT
5' ss Score
9.11
Exon sequences
Seq C1 exon
AGATGGTATTTCTGGGCCAACATATTGGTGATTATCCCGGCATTTGCCAGGGCAAAACACGGATACCCGGTGTCAACGATGGGGAAGAATTTGAGTTAACTGAC
Seq A exon
AGATGTGCTTCCTCTCCGATAACATTTACGACTACTATAACGTATCCCAGGGTAAAGTAACTGTACCCAACATGGATGACGGTGAGGAATTCCAGCTTGCAGAT
Seq C2 exon
CAAGCCTTCGACATCTTGGGCTTCACCAAGCAGGAGAAGGAGGACGTGTACAGGATCACCGCCGCTGTCATGCACATGGGTGGCATGAAGTTCAAGCAACGTGGTCGCGAGGAGCAGGCTGAGCAGGACGGCGAGGAGGAGGGTGGCCGTGTGTCGAAGCTGTTCGGTTGCGATACCGCCGAGCTGTACAAGAACTTGCTGAAGCCCCGCATCAAGGTCGGCAACGAGTTCGTCACCCAGGGCCGTAACGTCCAGCAGGTCACCAACTCGATCGGTGCCCTCTGCAAGGGTGTGTTCGATCGTCTGTTCAAGTGGCTGGTGAAGAAGTGTAACGAGACTCTGGATACCCAGCAGAAGCGTCAGCACTTCATTGGTGTACTGGATATTGCTGGTTTTGAGATCTTCGAG
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0264695-'20-15,'20-14,21-15
Average complexity
C3
Mappability confidence:
100%=100=100%
Protein Impact
In the CDS, with uncertain impact
No structure available
Features
Disorder rate (Iupred):
C1=0.057 A=0.029 C2=0.098
Domain overlap (PFAM):
C1:
PF0006316=Myosin_head=FE(5.0=100)
A:
PF0006316=Myosin_head=FE(5.0=100)
C2:
PF0006316=Myosin_head=FE(19.9=100)
Main Inclusion Isoform:
FBpp0080454
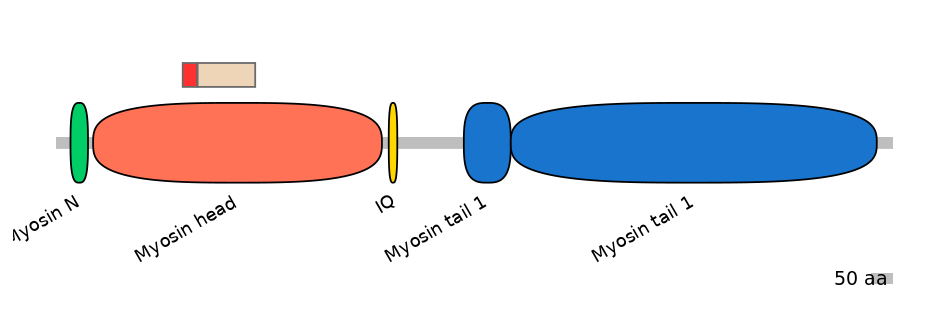
Main Skipping Isoform:
FBpp0312323
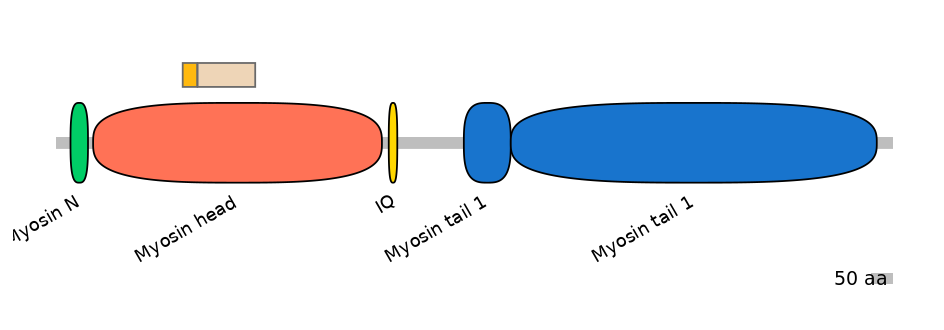
Other Inclusion Isoforms:
FBpp0080462, FBpp0080463, FBpp0080464, FBpp0291043
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GATACCCGGTGTCAACGATGG
R:
GGGGCTTCAGCAAGTTCTTGT
Band lengths:
251-355
Functional annotations
There are 3 annotated functions for this event
PMID: 16154586
This event
Exon 7 domain modulates myosin ATPase activity but has no effect on actin filament velocity. Transgenic flies expressing the indirect flight isoform-exon 7a (DmeEX0005109) have normal indirect flight muscle structure, and flight and jump ability. However, expression of the embryonic-exon 7d (DmeEX0005111) chimeric isoform yields flightless flies that show improvements in both the structural stability of the indirect flight muscle and in locomotor abilities as compared to flies expressing the embryonic isoform.
PMID: 16399836
This event
IFM expressing EMB with the exon 7a (DmeEX0005109) domain replaced by the IFM specific exon 7d (DmeEX0005111) domain (EMB-7d) exhibited 3.2-fold greater maximum oscillatory power (Pmax) and 1.5-fold greater optimal frequency of power generation (fmax) versus fibers expressing EMB. In contrast, IFM expressing IFI with the exon 7d region replaced by the EMB exon 7a region (IFI-7a), showed no change in Pmax, fmax, step response, or isometric muscle properties compared to native IFI fibers. A slight decrement in IFI-7a flight ability was observed, suggesting a negative influence of the increased ATPase rate on Drosophila locomotion, perhaps due to energy supply constraints. These results show that exon 7 plays a substantial role in establishing fiber speed and flight performance, and that the limiting step that sets ATPase rate in Drosophila myosin has little to no direct influence in setting fmax for fast muscle fiber types.
PMID: 17379245
This event
Two chimeras exchanged the entire exon 7 domain between the indirect flight muscle (IFI, normally containing exon 7d) and embryonic body wall muscle (EMB, normally containing exon 7a) isoforms to create IFI-7a and EMB-7d. The second two chimeras replaced each half of the exon 7a domain in EMB with the corresponding portion of exon 7d to create EMB-7a/7d and EMB-7d/7a. Transient kinetic studies of the motor domain from these myosin isoforms revealed changes in several kinetic parameters between the IFI or EMB isoforms and the chimeras. Of significance were changes in nucleotide binding, which differed in the presence and absence of actin, consistent with a model in which the exon 7 domain is part of the communication pathway between the nucleotide and actin-binding sites. Homology models of the structures suggest how the exon 7 domain might modulate this pathway.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)