DmeEX0005118 @ dm6
Exon Skipping
Gene
FBgn0264695 | Mhc
Description
The gene Myosin heavy chain is referred to in FlyBase by the symbol DmelMhc (CG17927, FBgn0264695). It is a protein_coding_gene from Dmel. It has 22 annotated transcripts and 22 polypeptides (20 unique). Gene sequence location is 2L:16766566..16788766. Its molecular function is described by 6 unique terms, many of which group under: nucleoside-triphosphatase activity; catalytic activity; hydrolase activity, acting on acid anhydrides; drug binding; actin filament binding. It is involved in the biological process described with 15 unique terms, many of which group under: epithelial cell development; adult somatic muscle development; regulation of biological quality; reproductive process; protein stabilization. 161 alleles are reported. The phenotypes of these alleles manifest in: somatic cell of ovariole; muscle attachment site; embryonic/larval heart; adult visceral muscle; germline cell. The phenotypic classes of alleles include: sterile; fertile; phenotype; increased mortality during development.
Coordinates
chr2L:16782775-16785841:+
Coord C1 exon
chr2L:16782775-16783661
Coord A exon
chr2L:16783833-16783911
Coord C2 exon
chr2L:16784666-16785841
Length
79 bp
Sequences
Splice sites
3' ss Seq
TTACCCATTTGCTAATCCAGGGC
3' ss Score
6.39
5' ss Seq
AAGGTAATA
5' ss Score
8.49
Exon sequences
Seq C1 exon
AAGTCCAAGCGCAAGGTTGAGGGCGACCTCAAGCTCACCCAGGAGGCTGTTGCCGATCTGGAGCGCAACAAGAAGGAGCTCGAGCAGACCATCCAGCGCAAGGACAAGGAGCTGTCCTCCATCACCGCCAAGCTCGAGGACGAGCAGGTCGTTGTTCTGAAGCACCAGCGCCAGATCAAGGAACTGCAGGCCCGCATCGAGGAGCTCGAGGAAGAGGTCGAGGCTGAGCGCCAGGCCCGCGCCAAGGCTGAGAAGCAGCGCGCCGATCTGGCCCGCGAACTCGAGGAATTGGGCGAGCGTCTGGAGGAGGCTGGCGGTGCCACCTCTGCCCAGATTGAGCTCAACAAGAAGCGTGAGGCTGAGTTGAGCAAACTGCGTCGCGATCTTGAGGAGGCCAACATCCAGCACGAGTCCACCCTGGCTAACCTGCGCAAGAAGCACAACGATGCCGTCGCCGAGATGGCCGAGCAGGTTGATCAGCTCAACAAGCTGAAGGCTAA
Seq A exon
GGCTGAACACGATCGCCAGACTTGCCACAACGAGCTGAATCAGACTCGTACCGCCTGCGATCAGCTGGGTCGCGATAAG
Seq C2 exon
GCTGCCCAGGAGAAGATCGCCAAGCAGCTGCAGCACACCCTCAACGAGGTGCAGTCGAAACTGGACGAGACCAACAGGACTCTGAACGACTTCGATGCCAGCAAGAAGAAGCTGTCCATTGAGAACTCCGATCTGCTCCGCCAGCTGGAGGAGGCCGAGTCCCAGGTGTCTCAGCTGTCCAAGATCAAGATCTCTCTGACCACCCAGTTGGAGGACACCAAGCGTCTGGCCGACGAGGAGTCGCGCGAGCGTGCCACCCTTTTGGGCAAGTTCCGCAACTTGGAGCACGACCTGGACAATCTGCGCGAGCAGGTTGAGGAGGAGGCTGAGGGCAAGGCCGATCTGCAGCGCCAGCTGAGCAAGGCCAACGCTGAGGCCCAGGTGTGGCGCAGCAAGTACGAGTCCGATGGCGTTGCCCGCTCTGAGGAGCTGGAGGAAGCCAAGAGGAAGCTGCAGGCCCGTTTGGCCGAGGCCGAGGAGACCATCGAGTCCCTCAACCA
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0264695_MULTIEX2-1/2=C1-C2
Average complexity
ME(1-2[100=100])
Mappability confidence:
100%=100=100%
Protein Impact
In the CDS, with uncertain impact
No structure available
Features
Disorder rate (Iupred):
C1=0.781 A=0.963 C2=0.391
Domain overlap (PFAM):
C1:
PF131661=AAA_13=PD(87.9=58.8),PF0157614=Myosin_tail_1=WD(100=38.2),PF045827=Reo_sigmaC=WD(100=47.0),PF0157614=Myosin_tail_1=PU(17.2=50.0),PF0076914=ERM=PU(26.1=8.1)
A:
PF0157614=Myosin_tail_1=FE(3.0=100)
C2:
PF0157614=Myosin_tail_1=FE(45.5=100),PF0076914=ERM=PD(43.5=10.2),PF0003816=Filament=WD(100=74.2),PF0076914=ERM=WD(100=32.7),PF131661=AAA_13=PU(83.4=38.5)
Main Inclusion Isoform:
FBpp0080454
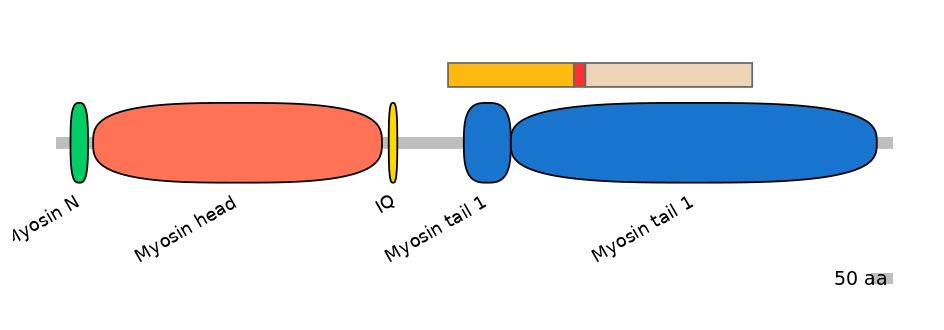
Main Skipping Isoform:
FBpp0080452
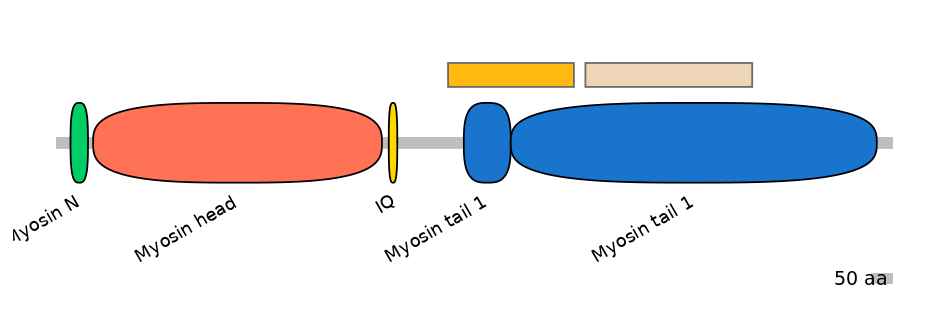
Other Inclusion Isoforms:
FBpp0080462, FBpp0080463, FBpp0080464, FBpp0291043, FBpp0298829, FBpp0312323
Other Skipping Isoforms:
FBpp0080453, FBpp0080455, FBpp0080456, FBpp0080457, FBpp0080458, FBpp0080459, FBpp0080460, FBpp0291041, FBpp0291042, FBpp0298827, FBpp0298828, FBpp0298830, FBpp0298831
Associated events
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CGTGAGGCTGAGTTGAGCAAA
R:
CTGCTTGGCGATCTTCTCCTG
Band lengths:
176-255
Functional annotations
There are 2 annotated functions for this event
PMID: 17316684
This event
A portion of the Drosophila melanogaster MHC hinge is encoded by mutually exclusive alternative exons 15a and 15b, the use of which correlates with fast (hinge A) or slow (hinge B) muscle physiological properties. To test the functional significance of alternative hinge regions, the authors constructed transgenic fly lines in which fast muscle isovariant hinge A was switched for slow muscle hinge B in the MHC isoforms of indirect flight and jump muscles. Substitution of the slow muscle hinge B impaired flight ability, increased sarcomere lengths by approximately 13% and resulted in minor disruption to indirect flight muscle sarcomeric structure compared with a transgenic control. With age, residual flight ability decreased rapidly and myofibrils developed peripheral defects. Computational analysis indicates that hinge B has a greater coiled-coil propensity and thus reduced flexibility compared to hinge A. Intriguingly, the MHC rod with hinge B was approximately 5 nm longer than myosin with hinge A, consistent with the more rigid coiled-coil conformation predicted for hinge B. This study demonstrates that hinge B cannot functionally substitute for hinge A in fast muscle types, likely as a result of differences in the molecular structure of the rod, subtle changes in myofibril structure and decreased ability to maintain sarcomere structure in indirect flight muscle myofibrils. Thus, alternative hinges are important in dictating the distinct functional properties of myosin isoforms and the muscles in which they are expressed.
PMID: 19450484
This event
The subfragment 2/light meromyosin hinge region has been proposed to significantly contribute to muscle contraction force and/or speed. Transgenic replacement of the endogenous fast muscle isovariant hinge A (exon 15a) in Drosophila melanogaster indirect flight muscle with the slow muscle hinge B (exon 15b) allows examination of the structural and functional changes when only this region of the myosin molecule is different. Hinge B was previously shown to increase myosin rod length, increase A-band and sarcomere length, and decrease flight performance compared to hinge A. The authors applied additional measures to these transgenic lines to further evaluate the consequences of modifying this hinge region. Structurally, the longer A-band and sarcomere lengths found in the hinge B myofibrils appear to be due to the longitudinal addition of myosin heads. Functionally, hinge B, although a significant distance from the myosin catalytic domain, alters myosin kinetics in a manner consistent with this region increasing myosin rod length. These structural and functional changes combine to decrease whole fly wing-beat frequency and flight performance.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)