DmeINT0019511 @ dm6
Intron Retention
Gene
FBgn0265296 | Dscam2
Description
The gene Down syndrome cell adhesion molecule 2 is referred to in FlyBase by the symbol DmelDscam2 (CG42256, FBgn0265296). It is a protein_coding_gene from Dmel. It has 16 annotated transcripts and 16 polypeptides (15 unique). Gene sequence location is 3L:7159199..7189931. Its molecular function is described by: cell-cell adhesion mediator activity; identical protein binding. It is involved in the biological process described with: homophilic cell adhesion via plasma membrane adhesion molecules; neuron projection morphogenesis; dendrite self-avoidance; axon guidance. 26 alleles are reported. The phenotypes of these alleles manifest in: adult external mesothorax; mesothoracic segment; thoracic segment; adult mesothoracic segment; dorsal mesothorax. The phenotypic classes of alleles include: neuroanatomy defective; size defective; viable; partially lethal - majority die; body color defective.
Coordinates
chr3L:7179723-7180586:-
Coord C1 exon
chr3L:7180138-7180586
Coord A exon
chr3L:7179868-7180137
Coord C2 exon
chr3L:7179723-7179867
Length
270 bp
Sequences
Splice sites
5' ss Seq
GTGGTGAGT
5' ss Score
8.95
3' ss Seq
CACCACATTCTGCTCCCCAGATG
3' ss Score
7.19
Exon sequences
Seq C1 exon
CTGGTTCACCCACAATGGAGCCGGACCCCTGCCCGTGCTGAGTGGCCCACGTGTCCGACTGCTTGGCCCCATCCTGGCCATTGAGGCTGTGACTGGCGAGGATTCCGGCGTCTACAAGTGCACGGCCGGCAATGTGGGCGGCGAAGCTAGTGCCGAGCTCCGGCTCACGGTGGCCACGCCCATCCAAGTGGAAATCTCACCGAACGTTCTCAGCGTTCACATGGGCGGCACCGCGGAGTTCCGGTGCCTGGTCACCAGCAACGGCAGTCCCGTGGGCATGCAGAACATCCTGTGGTACAAAGACGGCCGGCAGCTGCCCTCATCGGGCCGCGTCGAGGACACCCTGGTTGTGCCTCGCGTGAGTCGCGAGAACCGCGGTATGTACCAGTGCGTGGTGCGACGACCCGAAGGCGATACGTTCCAGGCCACCGCCGAGCTGCAACTGGGTG
Seq A exon
GTGAGTCCATCTAAGCTGTAAATATTAGTGGGCAAAAGGAATTTAAATTTATGATTTGAACCATAACTCTATTGTGAAGTAAAATCTTTAATATCTATTCCCATTTTTATTGTAAGCCGCTTATTTTAATTCCTTCTCCGACTTACAAACAATTCTATTCCCCACTTTGAAATCCTTTATGTATTAGAGCATCTAATCCCCCTCCCGGATTGGACCAGCTTATGGTTAAATTATTAATGTGACCTCTTGCCACCACATTCTGCTCCCCAG
Seq C2 exon
ATGCGCCGCCCGTGTTACTCTACAGCTTTATCGAGCAGACCCTGCAGCCAGGACCAGCTGTAAGCCTCAAGTGCTCCGCTGCCGGCAATCCTACGCCGCAAATTTCATGGACACTGGACGGATTTCCGCTGCCATCAAACGGAAG
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0265296:FBtr0299546:6
Average complexity
IR
Mappability confidence:
NA
Protein Impact
ORF disruption upon sequence inclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.046 A=NA C2=0.045
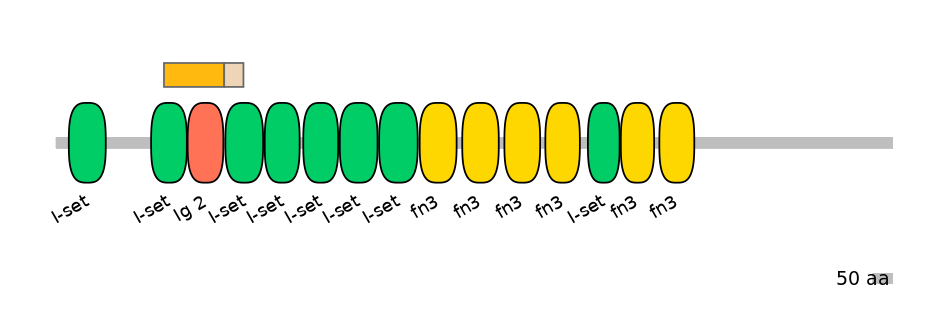
Domain overlap (PFAM):
C1:
PF0767911=I-set=PD(63.3=37.7),PF138951=Ig_2=WD(100=59.6)
A:
NA
C2:
PF0767911=I-set=PU(47.4=91.8)
Main Inclusion Isoform:
NA
Main Skipping Isoform:
FBpp0303367
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
FBpp0288821, FBpp0288822, FBpp0288823, FBpp0288824, FBpp0288825, FBpp0288826, FBpp0288827, FBpp0291333, FBpp0291334, FBpp0303368, FBpp0303369, FBpp0308278, FBpp0308279, FBpp0312398
Associated events
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GCATGCAGAACATCCTGTGGT
R:
GGAAATCCGTCCAGTGTCCAT
Band lengths:
301-571
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)