DreEX6054856 @ danRer10
Exon Skipping
Gene
ENSDARG00000022000 | traf3
Description
TNF receptor-associated factor 3 [Source:ZFIN;Acc:ZDB-GENE-040801-257]
Coordinates
chr17:29215725-29218185:-
Coord C1 exon
chr17:29218105-29218185
Coord A exon
chr17:29217903-29217977
Coord C2 exon
chr17:29215725-29215817
Length
75 bp
Sequences
Splice sites
3' ss Seq
TGTGGATGTGTCTCTCTTAGCTG
3' ss Score
6.65
5' ss Seq
AAGGTGACT
5' ss Score
6.64
Exon sequences
Seq C1 exon
AAACACAAAGAAACCGTCTGTCCTGCATTTCCTGTAGCATGTCCTAATCATTGCTCGTTTTCTTCTATTCTAAGAAGTGAG
Seq A exon
CTGTCAAGTCACCAGCATGACTGTCCAAAGGCTCAGGTGACCTGTTCCTTCATTCGCTACGGATGCTCCTACAAG
Seq C2 exon
GGGCTGAATCAAGAGATGAGGGAGCATGAGTCCAGTTTTGCATCTGAGCATCTAAGAATGATGGCAGTCAGAAACACCACACTAGAAGCCAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000022000-'9-8,'9-7,10-8=AN
Average complexity
A_S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.258
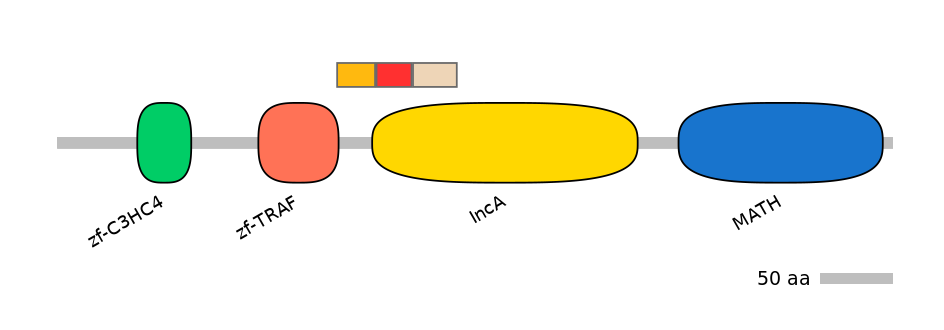
Domain overlap (PFAM):
C1:
PF0217613=zf-TRAF=PD(1.8=3.7),PF073947=DUF1501=FE(9.8=100),PF041569=IncA=PU(1.1=7.4),PF086146=ATG16=PU(0.1=0.0)
A:
PF073947=DUF1501=FE(9.0=100),PF041569=IncA=FE(13.1=100),PF086146=ATG16=FE(15.9=100),PF130941=CENP-Q=PU(6.7=44.0),PF061607=EzrA=PU(1.6=8.0)
C2:
PF073947=DUF1501=FE(11.3=100),PF041569=IncA=FE(16.4=100),PF086146=ATG16=FE(19.9=100),PF130941=CENP-Q=FE(18.3=100),PF061607=EzrA=FE(23.6=100),PF041027=SlyX=PU(6.2=12.9)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CAAAGAAACCGTCTGTCCTGC
R:
TGGCTTCTAGTGTGGTGTTTCTG
Band lengths:
167-242
Functional annotations
There are 1 annotated functions for this event
PMID: 24671418
The noncanonical nuclear factor _B (ncNF_B) pathway regulates the expression of chemokines required for secondary lymphoid organ formation and thus plays a pivotal role in adaptive immunity. Whereas ncNF_B signaling has been well described in stromal cells and B cells, its role and regulation in T cells remain largely unexplored. ncNF_B activity critically depends on the upstream NF_B-inducing kinase (NIK). NIK expression is negatively regulated by the full-length isoform of TNF receptor-associated factor 3 (Traf3) as formation of a NIK-Traf3-Traf2 complex targets NIK for degradation. The authors show that T cell-specific and activation-dependent alternative splicing generates a Traf3 isoform lacking exon 8 (Traf3DE8) (HsaEX0066805) that, in contrast to the full-length protein, activates ncNF_B signaling. Traf3DE8 disrupts the NIK-Traf3-Traf2 complex and allows accumulation of NIK to initiate ncNF_B signaling in activated T cells.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]