DreEX6064230 @ danRer10
Exon Skipping
Gene
ENSDARG00000058546 | si:dkey-30c15.12
Description
si:dkey-30c15.12 [Source:ZFIN;Acc:ZDB-GENE-060503-920]
Coordinates
chr18:7453261-7456175:-
Coord C1 exon
chr18:7455931-7456175
Coord A exon
chr18:7455649-7455791
Coord C2 exon
chr18:7453261-7453485
Length
143 bp
Sequences
Splice sites
3' ss Seq
TAAATATGTTCTAATGTCAGGTA
3' ss Score
6.54
5' ss Seq
ATGGTAATT
5' ss Score
6.49
Exon sequences
Seq C1 exon
GCTCTGGACTTTGAACAGGCCAACAGTTTCTCTCTGCTGATAATGGTAGAGAACGAGGCTCCTTTTGCGCTTCCTTTCCGCACCTCCACTGCTACCGTCACCATCACAGTCCTGGATGTAAATGAACCTCCAGTTTTTGATCTTCCAGAGAAACAGATCATAATTTATGAAGACCAATCTGTGGGCAGCATGATCTCCAAATATACAGCCACTGACCCAGACACTGATAGAGCACAGAAAATAAG
Seq A exon
GTATAAGCTGAGAGGAGACATTGCAAACTGGCTGGAGATCACAGAAGACACTGGACACATCACAATCAGCAGTTCGCTGGACAGAGAAGCTAAGTTCATAAGAGATGGGCAGTACAAGGTCTTAGTACTCGCATTCGATGATG
Seq C2 exon
ATGATGAACCGGCCACAGGCACAGGCACATTCATGATTAAGCTGTTGGATGTTAATGACAACCCTCCTGTTTTGGAGCAGCACAAAGTCCAGATGTGTTCAACAGAGCCAAAACCTGTCCGTCTGACCATCACAGATGCAGATGAGCCAGAAAACGGCCCACCGTTTACCACAGAGCTCCATGAAGAGTTTAAAAGCAACTGGACGATCTCCAGCGACTCCAACA
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000058546-'6-7,'6-6,7-7=AN
Average complexity
A_S
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.048 A=0.000 C2=0.373
Domain overlap (PFAM):
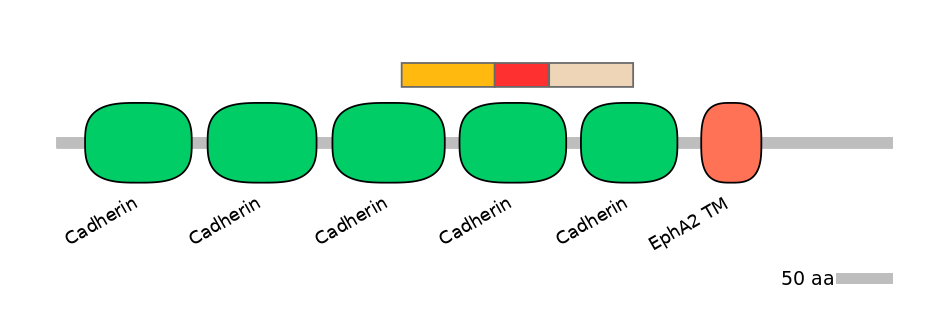
C1:
PF0002812=Cadherin=PD(38.0=45.8),PF053416=DUF708=WD(100=96.4),PF0002812=Cadherin=PU(32.6=37.3)
A:
PF0002812=Cadherin=FE(49.5=100)
C2:
PF0002812=Cadherin=PD(15.8=20.0),PF0002812=Cadherin=PU(53.5=61.3)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Rat
(rn6)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CCAATCTGTGGGCAGCATGAT
R:
GAGCTCTGTGGTAAACGGTGG
Band lengths:
251-394
Functional annotations
There are 2 annotated functions for this event
PMID: 19745069
The study used a microarray technique to screen for genes that up-regulate their RNA signal upon nonsense-mediated decay pathway blockade in chronic lymphocytic leukemia (CLL) specimens and identified an E-cadherin transcript with premature termination codon (PTC). Sequencing revealed an aberrant E-cadherin transcript lacking exon 11 (HsaEX0014149), resulting in a frameshift and PTC. The aberrant E-cadherin transcript was also identified in normal B cells, but occurred at a much lower level compared with CLL cells. In CLL specimens, E-cadherin expression was depressed more than 50% in 62% cases (relative to normal B cells). By real-time polymerase chain reaction analysis, the relative amounts of wild-type transcript inversely correlated with amounts of aberrant transcript (P = .018). Ectopic expression of E-cadherin in CLL specimens containing high amounts of aberrant transcript resulted in down-regulation of the wnt?beta-catenin pathway reporter, a pathway known to be up-regulated in CLL. These data point to a novel mechanism of E-cadherin gene inactivation, with CLL cells displaying a higher proportion of aberrant nonfunctional transcripts and resulting up-regulation of the wnt?beta-catenin pathway.
PMID: 21764905
[Disease and regulation]. The study analyzed the role of E-cadherin missplicing as a mechanism of its downregulation by analyzing a misspliced E-cadherin transcript that lacks exon 11 (HsaEX0014149) of this gene. This results in a frameshift and a premature termination codon that targets this transcript for degradation. Tumor tissues, including breast (20%, n = 9), prostate (30%, n = 9) and head and neck (75%, n = 8) cancer, express the exon 11-skipped transcripts (vs. nonmalignant controls) and its levels inversely correlate with E-cadherin expression. This is a novel mechanism of E-cadherin downregulation by missplicing in tumor cells, which is observed in highly prevalent human tumors. In the head and neck cancer model, nontumorigenic keratinocytes express exon 11-skipped splice product two- to sixfold lower than the head and neck tumor cell lines. Mechanistic studies reveal that SFRS2 (SC35), a splicing factor, as one of the regulators that increases missplicing and downregulates E-cadherin expression. Furthermore, this splicing factor was found to be overexpressed in 5 of 7 head and neck cell lines and primary head and neck tumors. Also, methylation of E-cadherin gene acts as a regulator of this aberrant splicing process. In 2 head and neck cell lines, wild-type transcript expression increased 16- to 25-folds, whereas the percentage of exon 11-skipped transcripts in both the cell lines decreased five- to 30-folds when cells were treated with a hypomethylating agent, azacytidine. These findings reveal that promoter methylation and an upregulated splicing factor (SFRS2) are involved in the E-cadherin missplicing in tumors.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]